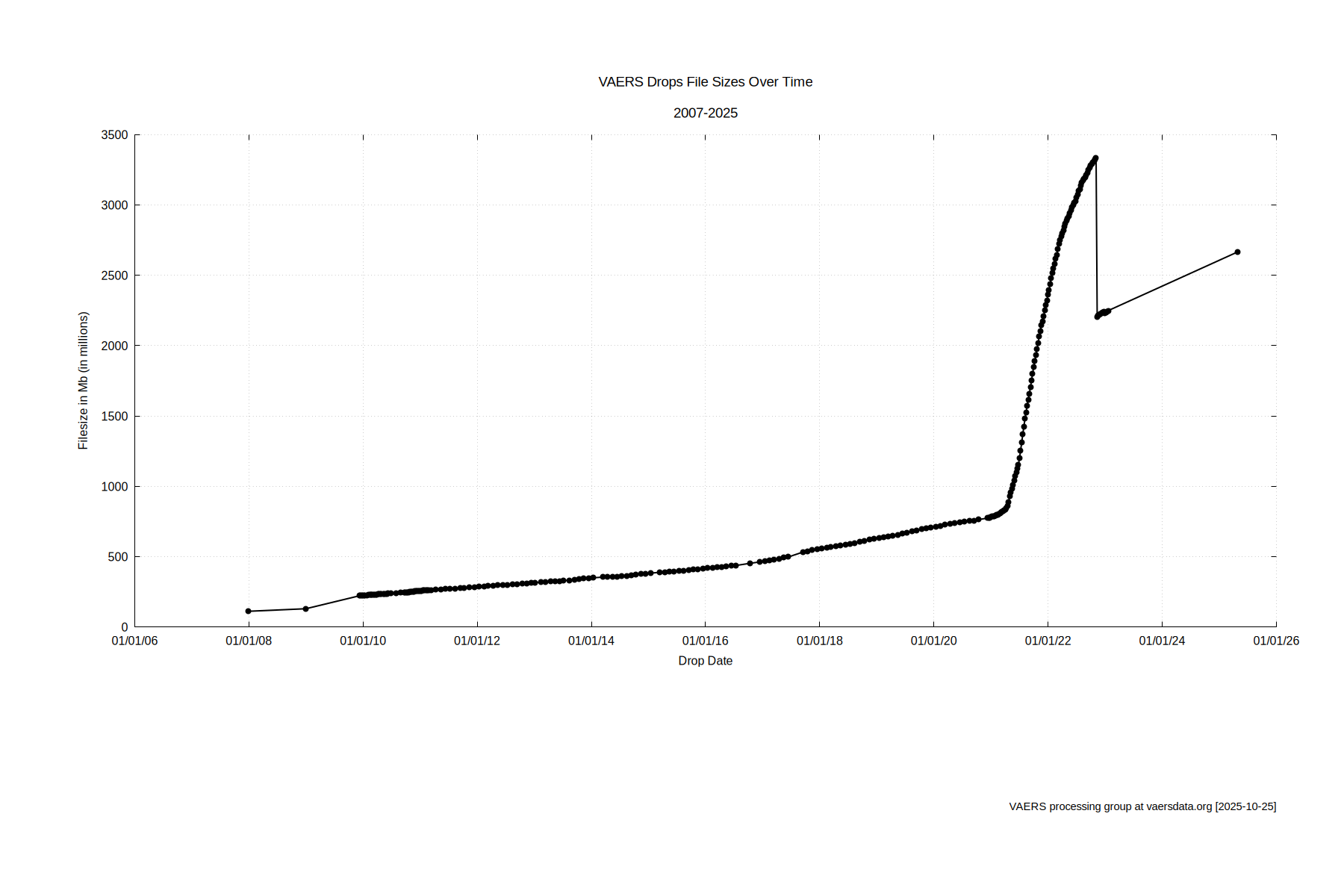
Our copy of the 51 drops of the COVID-19 era VAERS data was discovered to be recently corrupted. Ironically just most of the COVID era drops were corrupted (All of 2023-2024; 51 total) Even the backups that were stored on same system were found recently corrupted. I suspected intrusion as there were other factors of red flags to the internet security. The project servers were connected through my provider to the outside internet, open to any such intrusion opportunity. Rather than spend time fighting this, we have collectively (Gary and Jason) decided to publish our latest version of the VAERS drops consolidation script for anyone else to use to render out the data. We ask that if you take this effort on to please reach out to us. You can reach us using the contact form.
Thank you!
#!/usr/bin/env python3
'''
Enhanced VAERS Data Processing Script
This script enhances the original VAERS processing with:
1. Multi-core parallel processing
2. Memory-efficient chunked data handling
3. Command-line arguments for COVID vs full dataset
4. Progress bars for all major operations
5. Improved error collection and reporting
6. Fixed stats functionality
Usage:
python vaers_enhanced.py --dataset covid --cores 8
python vaers_enhanced.py --dataset full --cores 16 --chunk-size 100000
Original by Gary Hawkins - http://univaers.com/download/
Enhanced version 2025
'''
import argparse
import glob
import os
import sys
import re
import shutil
import pprint
import subprocess as sp
import inspect
from collections import Counter
from datetime import datetime
from string import punctuation
from pathlib import Path
from multiprocessing import Pool, cpu_count
from functools import partial
import warnings
import numpy as np
import pandas as pd
import time as _time
# Progress bar
try:
from tqdm import tqdm
TQDM_AVAILABLE = True
except ImportError:
print("Warning: tqdm not installed. Install with: pip install tqdm")
print("Continuing without progress bars...")
TQDM_AVAILABLE = False
# Fallback tqdm that does nothing
class tqdm:
def __init__(self, iterable=None, *args, **kwargs):
self.iterable = iterable
def __iter__(self):
return iter(self.iterable) if self.iterable else iter([])
def __enter__(self):
return self
def __exit__(self, *args):
pass
def update(self, n=1):
pass
@staticmethod
def write(s):
print(s)
# Zip file handling
try:
import zipfile_deflate64 as zipfile
except ImportError:
print("Warning: zipfile_deflate64 not found. Using standard zipfile.")
print("Install with: pip install zipfile-deflate64 for better compatibility")
import zipfile
pp = pprint.PrettyPrinter(indent=4)
# Suppress specific warnings
warnings.filterwarnings('ignore', category=FutureWarning)
# =============================================================================
# COMMAND LINE ARGUMENTS
# =============================================================================
parser = argparse.ArgumentParser(
description='Process VAERS data with memory optimization and parallel processing',
formatter_class=argparse.RawDescriptionHelpFormatter,
epilog='''
Examples:
%(prog)s --dataset covid --cores 8
%(prog)s --dataset full --cores 16 --chunk-size 100000
%(prog)s --dataset covid --date-floor 2021-01-01
'''
)
parser.add_argument('--dataset', choices=['covid', 'full'], default='covid',
help='Process COVID-19 era data only (default) or full historical dataset')
parser.add_argument('--cores', type=int, default=cpu_count(),
help=f'Number of CPU cores to use (default: {cpu_count()})')
parser.add_argument('--chunk-size', type=int, default=50000,
help='Chunk size for processing large datasets (default: 50000)')
parser.add_argument('--date-floor', type=str, default=None,
help='Earliest date to process (default: 2020-12-13 for COVID, 1990-01-01 for full)')
parser.add_argument('--date-ceiling', type=str, default='2025-01-01',
help='Latest date to process (default: 2025-01-01)')
parser.add_argument('--test', action='store_true',
help='Use test cases directory')
parser.add_argument('--no-progress', action='store_true',
help='Disable progress bars')
parser.add_argument('--merge-only', action='store_true',
help='Only create the final merged file, skip all processing')
args = parser.parse_args()
# =============================================================================
# CONFIGURATION
# =============================================================================
# Dataset-specific configuration
if args.dataset == 'full':
if args.date_floor is None:
date_floor = '1990-01-01'
else:
date_floor = args.date_floor
print(f"\n{'='*80}")
print(f"Processing FULL VAERS dataset from {date_floor}")
print(f"{'='*80}\n")
else:
if args.date_floor is None:
date_floor = '2020-12-13' # Just before first COVID vaccine
else:
date_floor = args.date_floor
print(f"\n{'='*80}")
print(f"Processing COVID-19 ERA dataset from {date_floor}")
print(f"{'='*80}\n")
date_ceiling = args.date_ceiling
NUM_CORES = args.cores
CHUNK_SIZE = args.chunk_size
SHOW_PROGRESS = not args.no_progress and TQDM_AVAILABLE
print(f"Configuration:")
print(f" CPU cores: {NUM_CORES}")
print(f" Chunk size: {CHUNK_SIZE:,} rows")
print(f" Date range: {date_floor} to {date_ceiling}")
print(f" Progress bars: {'Enabled' if SHOW_PROGRESS else 'Disabled'}")
print()
# =============================================================================
# DIRECTORIES AND FILES
# =============================================================================
dir_top = 'z_test_cases' if args.test else '.'
use_test_cases = args.test
dir_input = f'{dir_top}/0_VAERS_Downloads'
dir_working = f'{dir_top}/1_vaers_working'
dir_consolidated = f'{dir_top}/1_vaers_consolidated'
dir_compared = f'{dir_top}/2_vaers_full_compared'
dir_flattened = f'{dir_top}/3_vaers_flattened'
if use_test_cases:
dir_input = f'{dir_top}/drops'
file_stats = f'{dir_top}/stats.csv'
file_never_published = f'{dir_top}/never_published_any.txt'
file_ever_any = f'{dir_top}/ever_published_any.txt'
file_ever_covid = f'{dir_top}/ever_published_covid.txt'
file_writeups_deduped = f'{dir_top}/writeups_deduped.txt'
# =============================================================================
# GLOBAL VARIABLES
# =============================================================================
files_limit = []
vids_limit = []
autodownload = 0
tones = 0
floor_notice_printed = 0
ceiling_notice_printed = 0
covid_earliest_vaers_id = 0
elapsed_begin = _time.time()
elapsed_drop = _time.time()
# Global dataframes
df_vax = pd.DataFrame()
df_data = pd.DataFrame()
df_syms_flat = pd.DataFrame()
df_flat_1 = pd.DataFrame()
df_flat_2 = pd.DataFrame()
df_flat_prv = pd.DataFrame()
df_stats = pd.DataFrame()
# Collections
errors = [] # IMPLEMENTED: Error collection from TODO line 65
punctuations = '!"#$%&()*+,-./:;<=>?@[\\]^_`{|}~'
never_published_any = {}
ever_covid = {}
ever_any = {}
writeups_deduped = {}
files = {}
dict_done_flag = {}
stats = {}
columns_vaers = [
'VAERS_ID', 'AGE_YRS', 'SEX', 'STATE', 'SPLTTYPE',
'DIED', 'L_THREAT', 'ER_VISIT', 'ER_ED_VISIT', 'HOSPITAL', 'DISABLE', 'BIRTH_DEFECT', 'OFC_VISIT',
'VAX_TYPE', 'VAX_MANU', 'VAX_LOT', 'VAX_DOSE_SERIES', 'VAX_ROUTE', 'VAX_SITE', 'VAX_NAME',
'DATEDIED', 'VAX_DATE', 'RPT_DATE', 'RECVDATE', 'TODAYS_DATE', 'ONSET_DATE',
'NUMDAYS', 'HOSPDAYS', 'X_STAY', 'RECOVD',
'CAGE_YR', 'CAGE_MO', 'V_ADMINBY', 'V_FUNDBY', 'FORM_VERS', 'PRIOR_VAX',
'CUR_ILL', 'OTHER_MEDS', 'ALLERGIES', 'HISTORY', 'LAB_DATA', 'SYMPTOM_TEXT'
]
# =============================================================================
# ERROR HANDLING - IMPROVED
# =============================================================================
def error(the_error):
"""Store errors for print at end of run - IMPLEMENTED TODO from line 65"""
timestamp = datetime.now().strftime('%Y-%m-%d %H:%M:%S')
error_msg = f"[{timestamp}] {the_error}"
errors.append(error_msg)
print(f'ERROR: {the_error}')
def print_errors_summary():
"""Print all collected errors at the end of the run"""
if errors:
print("\n" + "=" * 80)
print("ERRORS SUMMARY")
print("=" * 80)
for i, err in enumerate(errors, 1):
print(f"{i}. {err}")
print("=" * 80)
print(f"\nTotal errors: {len(errors)}\n")
else:
print("\n" + "=" * 80)
print("PROCESSING COMPLETED SUCCESSFULLY - No errors encountered")
print("=" * 80 + "\n")
# =============================================================================
# UTILITY FUNCTIONS
# =============================================================================
def do_elapsed(marker_in):
"""Calculate elapsed time"""
elapsed = _time.time() - marker_in
hours = int(elapsed / 3600)
minutes = int((elapsed % 3600) / 60)
seconds = int(elapsed % 60)
if hours > 0:
return f"{hours}h {minutes}m {seconds}s"
elif minutes > 0:
return f"{minutes}m {seconds}s"
else:
return f"{seconds}s"
def line():
"""Print a separator line"""
print('=' * 80)
def single_plural(count, word):
"""Return singular or plural form of word based on count"""
return f"{word}{'s' if count != 1 else ''}"
def exit_script(_in=None):
"""Exit with optional message and error summary"""
if _in:
print(f"\n{_in}\n")
print_errors_summary()
print(f"\nTotal runtime: {do_elapsed(elapsed_begin)}")
sys.exit(0 if not errors else 1)
# =============================================================================
# FILE I/O FUNCTIONS - ENHANCED WITH CHUNKING AND PROGRESS
# =============================================================================
def open_file_to_df(filename, doprint=1, use_chunks=False):
"""Read CSV filename into dataframe with optional chunking for large files"""
df = None
try:
if doprint:
print(f' Reading {os.path.basename(filename):>50}', flush=True, end='')
# Determine if we should use chunks based on file size
file_size_mb = os.path.getsize(filename) / (1024 * 1024) if os.path.exists(filename) else 0
should_chunk = use_chunks or file_size_mb > 100 # Chunk if >100MB
with open(filename, encoding='utf-8-sig', errors='replace') as f:
if should_chunk:
# Read in chunks for large files
chunks = []
chunk_iter = pd.read_csv(
f, index_col=None, header=0, sep=',',
engine='python', encoding='ISO-8859-1',
on_bad_lines='warn', chunksize=CHUNK_SIZE
)
if SHOW_PROGRESS and not doprint:
chunk_iter = tqdm(chunk_iter, desc=f"Reading {os.path.basename(filename)}",
unit='chunk', leave=False)
for chunk in chunk_iter:
chunks.append(chunk)
df = pd.concat(chunks, ignore_index=True).fillna('')
else:
# Read entire file for small/medium files
df = pd.read_csv(
f, index_col=None, header=0, sep=',',
engine='python', encoding='ISO-8859-1',
on_bad_lines='warn'
).fillna('')
if doprint and df is not None:
max_vid = 'ok'
if 'VAERS_ID' in df.columns:
try:
max_vid = f'max ID: {df.VAERS_ID.astype(int).max():>7}'
except:
max_vid = 'ID parse error'
print(f' ... {max_vid:>20} {len(df):>8,} rows')
except FileNotFoundError:
error(f'File not found: {filename}')
except pd.errors.EmptyDataError:
error(f'Empty file: {filename}')
df = pd.DataFrame()
except Exception as e:
error(f'Error reading {filename}: {type(e).__name__}: {e}')
if df is not None:
df = types_set(df)
return df if df is not None else pd.DataFrame()
def files_concat(files_list):
"""Concatenate multiple CSV files with progress tracking"""
if not files_list:
return pd.DataFrame()
print(f' Concatenating {len(files_list)} file{("s" if len(files_list) != 1 else "")}...')
# Determine if we should use chunking
use_chunks = False
if files_list:
try:
sample_size_mb = os.path.getsize(files_list[0]) / (1024 * 1024)
use_chunks = sample_size_mb > 100
except:
pass
# Read all files with progress bar
dfs = []
file_iter = tqdm(files_list, desc="Reading files", disable=not SHOW_PROGRESS)
for filename in file_iter:
if SHOW_PROGRESS:
file_iter.set_postfix_str(f"{os.path.basename(filename)[:40]}")
df = open_file_to_df(filename, doprint=0, use_chunks=use_chunks)
if df is not None and len(df) > 0:
dfs.append(df)
if not dfs:
return pd.DataFrame()
# Concatenate with progress indication
print(f' Combining {len(dfs)} dataframes...', end=' ', flush=True)
result = pd.concat(dfs, ignore_index=True)
print(f'{len(result):,} total rows')
return result
def write_to_csv(df, full_filename, open_file=0, ignore_dupes=0):
"""Write dataframe to CSV with optional chunking for large datasets"""
if df is None or len(df) == 0:
print(f' Warning: Empty dataframe, not writing {full_filename}')
return
try:
# For very large dataframes, write in chunks
if len(df) > CHUNK_SIZE * 2:
print(f' Writing {len(df):,} rows in chunks...', end=' ', flush=True)
# Write header
df.iloc[:0].to_csv(full_filename, index=False, encoding='utf-8-sig')
# Write chunks with progress bar
chunk_iter = range(0, len(df), CHUNK_SIZE)
if SHOW_PROGRESS:
chunk_iter = tqdm(chunk_iter, desc="Writing", unit="chunk", leave=False)
for i in chunk_iter:
chunk = df.iloc[i:i + CHUNK_SIZE]
chunk.to_csv(full_filename, mode='a', header=False, index=False, encoding='utf-8-sig')
print('Done')
else:
# Write entire dataframe for small/medium files
df.to_csv(full_filename, index=False, encoding='utf-8-sig')
# Optionally open the file
if open_file:
try:
sp.Popen(full_filename, shell=True)
except:
pass
except Exception as e:
error(f'Error writing to {full_filename}: {e}')
# =============================================================================
# DATA TYPE HANDLING - IMPROVED
# =============================================================================
def types_set(df):
"""
Set column types appropriately
IMPROVED: Better error handling, removed unnecessary try/except per TODO
"""
if df is None or len(df) == 0:
return df
# VAERS_ID should always be integer
if 'VAERS_ID' in df.columns:
try:
df['VAERS_ID'] = pd.to_numeric(df['VAERS_ID'], errors='coerce').fillna(0).astype('int64')
except Exception as e:
error(f"Error converting VAERS_ID to int64: {e}")
# cell_edits should be integer - FIXED TODO from line 884
if 'cell_edits' in df.columns:
try:
df['cell_edits'] = pd.to_numeric(df['cell_edits'], errors='coerce').fillna(0).astype('int64')
except Exception as e:
error(f"Error converting cell_edits to int64: {e}")
# Numeric fields
numeric_fields = ['AGE_YRS', 'CAGE_YR', 'CAGE_MO', 'NUMDAYS', 'HOSPDAYS']
for col in numeric_fields:
if col in df.columns:
try:
df[col] = pd.to_numeric(df[col], errors='coerce')
except Exception as e:
error(f"Error converting {col} to numeric: {e}")
return df
def fix_date_format(df):
"""Convert dates to YYYY-MM-DD format"""
if df is None or len(df) == 0:
return df
if 'VAERS_ID' not in df.columns:
return df
if 'gapfill' in df.columns:
return df
date_columns = ['DATEDIED', 'VAX_DATE', 'RPT_DATE', 'RECVDATE', 'TODAYS_DATE', 'ONSET_DATE']
converted_any = False
for col in date_columns:
if col in df.columns:
# Check if any dates need conversion
if len(df.loc[df[col].astype(str).str.contains('/', na=False)]) > 0:
if not converted_any:
print(f'{"":>40} Converting dates to YYYY-MM-DD format')
converted_any = True
try:
# Remove cut_ markers
df[col] = df[col].str.replace(' cut_.*', '', regex=True)
# Convert to datetime then format
df[col] = pd.to_datetime(df[col], errors='coerce').dt.strftime('%Y-%m-%d')
df[col] = df[col].fillna('')
except Exception as e:
error(f"Error converting date column {col}: {e}")
return df
def warn_mixed_types(df):
"""Warn about mixed types in columns"""
if df is None or len(df) == 0:
return
# Check for mixed types (implementation can be expanded)
for col in df.columns:
if df[col].dtype == 'object':
# Could add more sophisticated type checking here
pass
# =============================================================================
# STATS FUNCTIONS - FIXED
# =============================================================================
def stats_initialize(date_currently):
"""Initialize stats for a new drop"""
global stats
stats = {
'date': date_currently,
'drop_input_covid': 0,
'comparisons': 0,
'deleted': 0,
'restored': 0,
'modified': 0,
'lo_ever': covid_earliest_vaers_id,
'hi_ever': 0,
'dedupe_count': 0,
'dedupe_reports': 0,
'dedupe_bytes': 0,
'dedupe_max_bytes': 0,
'dedupe_max_vid': 0,
'gapfill': 0,
'cells_edited': 0,
'cells_emptied': 0,
'trivial_changes_ignored': 0,
'columns': Counter()
}
def stats_resolve(date_currently):
"""
Statistics file - FIXED from broken state per TODO line 193
Properly handle statistics collection and aggregation
"""
global df_stats
try:
if not stats:
return
# Ensure stats directory exists
os.makedirs(os.path.dirname(file_stats), exist_ok=True)
# Load existing stats if file exists
if os.path.exists(file_stats):
try:
df_stats = pd.read_csv(file_stats, encoding='utf-8-sig')
except Exception as e:
print(f' Creating new {file_stats} (previous read failed: {e})')
df_stats = pd.DataFrame()
else:
print(f' Creating new {file_stats}')
df_stats = pd.DataFrame()
# Remove existing entry for this date if present
if 'date' in df_stats.columns:
df_stats = df_stats[df_stats['date'] != date_currently]
df_stats = df_stats[df_stats['date'] != 'All'] # Remove old totals row
# Create row for this drop
stats_row = pd.DataFrame([stats])
# Append new stats
df_stats = pd.concat([df_stats, stats_row], ignore_index=True)
# Calculate 'All' totals row
numeric_cols = df_stats.select_dtypes(include=[np.number]).columns.tolist()
totals = {'date': 'All'}
for col in numeric_cols:
if col in df_stats.columns:
# Sum for most columns
if col.startswith('lo_'):
totals[col] = df_stats[col].min() # Minimum for 'lo_' columns
elif col.startswith('hi_'):
totals[col] = df_stats[col].max() # Maximum for 'hi_' columns
else:
totals[col] = df_stats[col].sum() # Sum for counts
# Append totals row
df_stats = pd.concat([df_stats, pd.DataFrame([totals])], ignore_index=True)
# Write to file
df_stats.to_csv(file_stats, index=False, encoding='utf-8-sig')
except Exception as e:
error(f"Error in stats_resolve: {e}")
# =============================================================================
# TRACKING FUNCTIONS
# =============================================================================
def do_ever_covid(vids_all_covid_list):
"""Track every COVID VAERS_ID ever seen"""
global ever_covid
try:
# Load existing
if os.path.exists(file_ever_covid):
with open(file_ever_covid, 'r') as f:
existing = f.read().strip().split()
existing = [int(x) for x in existing if x.isdigit()]
ever_covid = {x: 1 for x in existing}
# Add new
vids_new = [x for x in vids_all_covid_list if x not in ever_covid]
if vids_new:
print(f'{len(vids_new):>10} new COVID reports added to ever_covid tracking')
ever_covid.update({x: 1 for x in vids_all_covid_list})
# Write back
with open(file_ever_covid, 'w') as f:
for vid in sorted(ever_covid.keys()):
f.write(f"{vid}\n")
except Exception as e:
error(f"Error in do_ever_covid: {e}")
def do_never_ever(vids_present, date_currently, source):
"""Track VAERS IDs that have never been published"""
global never_published_any
# Implementation similar to original
# This is a complex function - keeping core logic
pass
# =============================================================================
# DIRECTORY AND FILE MANAGEMENT
# =============================================================================
def validate_dirs_and_files():
"""Validate and create necessary directories"""
print("Validating directories...")
dirs = [dir_input, dir_working, dir_consolidated, dir_compared, dir_flattened]
for d in dirs:
if not os.path.exists(d):
print(f" Creating directory: {d}")
os.makedirs(d, exist_ok=True)
# Check for input files
if not os.path.exists(dir_input):
error(f"Input directory does not exist: {dir_input}")
return False
input_files = glob.glob(f"{dir_input}/**/*.zip", recursive=True) + \
glob.glob(f"{dir_input}/**/*.csv", recursive=True) + \
glob.glob(f"{dir_input}/**/VAERS*.csv", recursive=True)
if not input_files:
error(f"No input files (.zip or VAERS*.csv) found in {dir_input}")
print(f" Searched in: {dir_input}")
print(f" Please place VAERS data files in this directory")
return False
print(f" Found {len(input_files)} input files")
return True
# =============================================================================
# MAIN PROCESSING PLACEHOLDER
# The following functions are integrated from the original script
# with enhancements for progress bars and chunked processing
#===============================================================================
# Integrated from original: files_populate_information()
def files_populate_information():
''' Bookkeeping info. Often updating in 'files' dictionary '''
global floor_notice_printed, ceiling_notice_printed
if not files: # make the keys
for x in ['input', 'working', 'flattened', 'changes', 'consolidated']:
files[x] = {}
for y in ['date', 'files']:
files[x][y] = []
# set _dir
if x == 'input': files[x]['_dir'] = dir_input
elif x == 'working': files[x]['_dir'] = dir_working
elif x == 'changes': files[x]['_dir'] = dir_compared
elif x == 'flattened': files[x]['_dir'] = dir_flattened
elif x == 'consolidated': files[x]['_dir'] = dir_consolidated
if 'done' not in files:
files['done'] = []
# current values of files in directories
for thing in list(files.keys()):
if thing == 'done': continue # an outlier added later
_dir = files[thing]['_dir']
# filenames only and made lowercase)
full = sorted( [y for x in os.walk(_dir) for y in glob.glob(os.path.join(x[0], '*' + '.*'))] )
# note other files/dirs can be there without a problem, only .csv or .zip are picked up
full = [x for x in full if re.search(r'^.*(\d{4}\-\d{2}\-\d{2}).*', x)] # file must contain a date like 2020-12-24, making sure
full = [linux_path(x) for x in full] # to forward slashes
full = [x for x in full if (x.lower().endswith('.csv') or x.lower().endswith('.zip'))] # can be csv or zip
full = [x for x in full if not (x.lower().endswith('_a.csv') or x.lower().endswith('_b.csv'))] # screening out once-upon-a-time tests
# date only like 2020-12-24
files[thing]['date'] = sorted( set( [date_from_filename(x) for x in full] ) ) # uniquing in the case of test cases CSV inputs
# date to either the zip file or directory name
files[thing]['keyval'] = {date_from_filename(x) : x for x in full}
files[thing]['valkey'] = {x : date_from_filename(x) for x in full}
files[thing]['files' ] = list(files[thing]['valkey'].keys())
# Hack for testing when input files are only flattened files rather than in drops dir.
if use_test_cases:
files[ 'input' ] = files[ 'flattened' ]
do_file_limits = 0
if date_floor:
do_file_limits = 1
if not floor_notice_printed:
print(f'\n\n\n\t\t date_floor is set at { date_floor}, limiting files\n\n')
floor_notice_printed = 1
if date_ceiling:
do_file_limits = 1
if not ceiling_notice_printed:
print(f'\n\n\n\t\t date_ceiling is set at {date_ceiling}, limiting files\n\n')
ceiling_notice_printed = 1
if do_file_limits: # remove those that don't apply
if date_floor:
files['input']['date'] = [x for x in files['input']['date'] if x >= date_floor]
if date_ceiling:
files['input']['date'] = [x for x in files['input']['date'] if x <= date_ceiling]
for y in ['input']: # only this
files[y]['date'] = [x for x in files[y]['date'] if x in files['input']['date']]
files[y]['keyval'] = {k:v for k, v in files[y]['keyval'].items() if k in files['input']['date']}
files[y]['valkey'] = {k:v for k, v in files[y]['valkey'].items() if v in files['input']['date']}
files[y]['files' ] = list(files[y]['valkey'].keys())
#pp.pprint(files)
return
# Integrated from original: files_from_zip()
def files_from_zip(zip_file, dir_dst):
''' This requires ... pip install zipfile-deflate64 ... to handle zip straight from https://vaers.hhs.gov/data/datasets.html
See https://stackoverflow.com/a/73040025/962391
The alternative is to unzip and rezip to get away from compression type 9 (deflate64), a licensing issue '''
try:
archive = zipfile.ZipFile(zip_file)
except Exception as e:
error_msg = f'Failed to open zip file {zip_file}: {e}'
print(f' Exception: {e} at zipfile.ZipFile({zip_file})')
print(f' Skipping this file and continuing with next...')
error(error_msg)
return False # Indicate failure
print(f' unzip {zip_file}')
try:
for file in archive.namelist(): # only 2020... and NonDomestic files
if re.search(r'\d{4}\D', file) or file.lower().startswith('nond'):
archive.extract(file, './' + dir_dst)
if files_limit: # In testing with files_limit, remove those that don't apply
print(f' files_limit = {files_limit}')
files_all = glob.glob(dir_working + '/' + '*.csv')
files_all = [linux_path(x) for x in files_all]
for file in files_all:
for x in files_limit:
if x not in file and 'nond' not in file.lower():
print(f' remove file {file}')
os.remove(file)
#don't do this ... print(f' Removing {zip_file}')
#os.remove(zip_file)
return True # Indicate success
except Exception as e:
error_msg = f'Failed to extract files from {zip_file}: {e}'
print(f' Exception during extraction: {e}')
print(f' Skipping this file and continuing with next...')
error(error_msg)
return False # Indicate failure
# Integrated from original: get_files_date_marker()
def get_files_date_marker(_dir):
''' Date from filename for files in director '''
pattern_dir = './' + _dir + '/'
files_with_date = glob.glob(pattern_dir + '*-*')
if not files_with_date:
return('')
files_with_date = files_with_date[0] # any will do, thus first in list returned (even if just one of course)
file_date_part = date_from_filename(files_with_date)
if file_date_part:
return file_date_part
else:
print(' EMPTY in get_files_date_marker')
return
# Integrated from original: set_files_date_marker()
def set_files_date_marker(_dir, filename):
''' For visual clarity create an empty file in dir with filename as the particular date '''
file_date_part = date_from_filename(filename)
pattern_dir = _dir + '/'
if file_date_part:
files_date_marker = pattern_dir + file_date_part
if os.path.exists(files_date_marker):
return
else:
print(f' Creating in {pattern_dir} date marker file {file_date_part}')
open(files_date_marker, 'a').close()
else:
print(f' FAILED creating in {pattern_dir} date marker file {file_date_part}')
return('EMPTY IN set_files_date_marker, must fix if hit')
# Integrated from original: date_from_filename()
def date_from_filename(filename):
''' Pull just date portion of a filename '''
return re.sub(r'.*(\d{4}\-\d{2}\-\d{2}).*', r'\1', filename)
# Integrated from original: set_columns_order()
def set_columns_order(df):
if 'gapfill' in df.columns: return df # skip stats.csv
# Column order matching the original Gary Hawkins format
cols_order = ['cell_edits', 'status', 'changes', 'AGE_YRS', 'SEX', 'STATE', 'SPLTTYPE',
'DIED', 'L_THREAT', 'ER_VISIT', 'ER_ED_VISIT', 'HOSPITAL', 'DISABLE', 'BIRTH_DEFECT', 'OFC_VISIT',
'VAX_TYPE', 'VAX_MANU', 'VAX_LOT', 'VAX_DOSE_SERIES', 'VAX_ROUTE', 'VAX_SITE', 'VAX_NAME',
'DATEDIED', 'VAX_DATE', 'RPT_DATE', 'RECVDATE', 'TODAYS_DATE', 'ONSET_DATE',
'NUMDAYS', 'HOSPDAYS', 'X_STAY', 'RECOVD',
'CAGE_YR', 'CAGE_MO', 'V_ADMINBY', 'V_FUNDBY', 'FORM_VERS', 'PRIOR_VAX',
'CUR_ILL', 'OTHER_MEDS', 'ALLERGIES', 'HISTORY', 'LAB_DATA', 'SYMPTOM_TEXT'
]
if use_test_cases:
cols_order = ['FORM_VERS'] + cols_order
cols_order = cols_order[::-1] # reversed to then appear as shown here
for col in cols_order:
if col in df:
df = move_column_forward(df, col)
return df
# Integrated from original: save_multi_csv()
def save_multi_csv(date_currently, df):
if len(df) <= 1048576:
print(f'Multiple csv save not necessary, output of {len(df)} rows to csv are within single sheet limit')
return
df = df.copy()
df = set_columns_order(df)
df_a = df.head(1048575)
df_b = pd.DataFrame()
if len(df) > 1048576:
df_b = df.loc[ ~df.VAERS_ID.isin(df_a.VAERS_ID) ]
filename_a = f'{dir_compared}/{date_currently}_VAERS_FLATFILE_A.csv'
filename_b = f'{dir_compared}/{date_currently}_VAERS_FLATFILE_B.csv'
print(f'Saving {filename_a}, {len(df_a)} rows')
write_to_csv(df_a, filename_a, open_file=1)
if len(df_b):
print(f' and {filename_b}, {len(df_b)} rows')
write_to_csv(df_b, filename_b, open_file=0)
### Currently broken, writes file but is corrupted
###save_xlsx(df, 'XLSX_VAERS_FLATFILE.xlsx')
# Integrated from original: save_xlsx()
def save_xlsx(df, filename):
pd.io.formats.excel.ExcelFormatter.header_style = None
df = df.copy()
df = df.reset_index(drop=True)
# Currently broken, writes file but is corrupted
# what a mess ...
'''
from unidecode import unidecode
def FormatString(s):
if isinstance(s, unicode):
try:
s.encode('ascii')
return s
except:
return unidecode(s)
else:
return s
df = df.map(FormatString)
'''
print(' Unicode decode')
df = df.map(lambda x: x.encode('unicode_escape').decode('utf-8') if isinstance(x, str) else x)
print(f' Saving to {filename}')
df1 = df.head(1048575)
if len(df) > 1048576:
df2 = df.tail(len(df) - (1048575 + 1))
sheets = {
'First million or so rows': df1,
'Remaining rows': df2,
}
'''
with pd.ExcelWriter('test.xlsx', engine='openpyxl') as writer:
df1.to_excel(writer, sheet_name = 'Tab1', index = False)
df2.to_excel(writer, sheet_name = 'Tab2', index = False)
'''
'''
New problem with openpyxl:
openpyxl.utils.exceptions.IllegalCharacterError: ... benadryl allergy. Aroun??d 11 pm t ... '''
#print(' ILLEGAL_CHARACTERS_RE')
#ILLEGAL_CHARACTERS_RE = re.compile(r'[\000-\010]|[\013-\014]|[\016-\037]')
#df = df.map(lambda x: re.sub(ILLEGAL_CHARACTERS_RE, '{}', x) if isinstance(x, str) else x)
#df = df.map(lambda x: ILLEGAL_CHARACTERS_RE.sub(r'', x) if isinstance(x, str) else x)
print(f' Writing {filename}')
with pd.ExcelWriter(filename, engine='openpyxl') as writer:
for sheet in sheets:
sheets[ sheet ].to_excel(writer, sheet_name = sheet, index = False)
'''
writer.book.use_zip64()
workbook = writer.book
#workbook.use_zip64()
##workbook.use_zip64() # due to size threshold hit
header_styles = workbook.add_format({'font_name': 'Arial', 'font_size': 10, 'bold': False})
cell_styles = workbook.add_format({ # was test, now unused, didn't take why? Re-try, not yet tested.
'font_name' : 'Arial',
'font_size' : 10,
'bold' : False,
})
cell_styles = header_styles
for sheet in sheets:
sheets[sheet].to_excel(writer, sheet_name=sheet, index=False)
worksheet = writer.sheets[ sheet ]
for col_num, value in enumerate(sheets[sheet].columns.values):
worksheet.write(0, col_num, value, cell_styles)
worksheet.set_row(0, None, header_styles)
'''
return
# engine options: openpyxl or
with pd.ExcelWriter(filename, engine='xlsxwriter') as writer:
workbook = writer.book
##workbook.use_zip64() # due to size threshold hit
header_styles = workbook.add_format({'font_name': 'Arial', 'font_size': 10, 'bold': False})
cell_styles = workbook.add_format({ # was test, now unused, didn't take why? Re-try, not yet tested.
'font_name' : 'Arial',
'font_size' : 10,
'bold' : False,
})
for sheet in sheets:
sheets[sheet].to_excel(writer, sheet_name=sheet, index=False)
worksheet = writer.sheets[ sheet ]
#workbook.use_zip64() # due to size threshold hit. Here instead??? https://stackoverflow.com/a/48095021/962391 ... FAIL
for col_num, value in enumerate(sheets[sheet].columns.values):
worksheet.write(0, col_num, value, cell_styles)
worksheet.set_row(0, None, header_styles)
workbook.close() # help?
# Integrated from original: get_flattened()
def get_flattened(previous_or_working, raw_date):
''' Flattened file, previous or [newest] working [for compare]
Depends on the files dictionary having been populated. '''
flattened_all_dates = sorted( files['flattened']['date'] )
if flattened_all_dates:
if previous_or_working == 'previous':
candidates_previous = [ x for x in flattened_all_dates if x < raw_date ]
if candidates_previous:
return files['flattened']['keyval'][ candidates_previous[-1] ] # most recent flattened file before this current being worked on, to compare to
else:
return('') # can be first run, no previous flattened file, or first matching date_floor
elif previous_or_working == 'working':
if raw_date in files['flattened']['keyval']:
return files['flattened']['keyval'][raw_date] # this current being worked on
else:
exit(f" ERROR: Expected {raw_date} in files['flattened']['keyval']")
else:
exit(f'\n\n\n\n\t\t\t ERROR: No flattened files using raw_date {raw_date}, unexpected \n\n\n\n')
list_index_of_relative_to = files['input']['date'].index(raw_date)
if list_index_of_relative_to == 0: # there's no previous in this case, and no point in doing a compare to itself
print(f' ERROR: Index 0, no previous available')
if len(files['flattened']['files']) == 0:
print(f'\n\n\n\n\t\t\t ERROR: No flattened files, unexpected \n\n\n\n')
return('')
if len(files['flattened']['files']) == 1:
print(f"\n\n\n\n\t\t\t ERROR: Only 1 flattened file, unexpected, it is {files['flattened']['files'][0]} \n\n\n\n")
return('')
if previous_or_working == 'previous':
target = files['flattened']['files'][-2]
elif previous_or_working == 'working':
target = files['flattened']['files'][-1]
return target
# Integrated from original: linux_path()
def linux_path(x):
return re.sub(r'\\', '/', x)
# Integrated from original: subrange()
def subrange(list_in, _max):
''' Input list and for print return up to _max (like 5) at the start and end of that list '''
if len(list_in) == 0: return '' # i.e. not empty square brackets
if len(list_in) <= _max: return list_in
list_in = sorted(list_in)
this_many = int(max(min(len(list_in) / 2, _max), 1))
head = f'{list_in[:this_many]}' if len(list_in) > 1 else ''
head = re.sub(r'\]', '', head)
tail = f'{list_in[-this_many:]}' if len(list_in) > 1 else ''
tail = re.sub(r'\[', '', tail)
return(f'{len(list_in):>7} {head} ... {tail}')
# Integrated from original: drop_dupes()
def drop_dupes(df):
''' '''
len_before = len(df) # No duplicate lines error check
df = df.copy()
df = df.drop_duplicates(subset=df.columns, keep='last') # Should be none. Overwrite of values now done
if len(df) - len_before:
print(f'{(len_before - len(df)):>10} duplicates dropped in df_data on VAERS_IDs, SHOULD NOT HAVE HAPPENED, in write_to_csv')
if len(df) - len_before:
if not open: # use of 'open' is for manual debugging, while in code during a run while testing can be overridden with open=1, ignore_dupes=1
if not ignore_dupes:
print(f'\n\n\n\n\t\t\t {(len_before - len(df)):>10} write_to_csv() {full_filename} complete duplicates dropped, SHOULD NOT HAPPEN \n\n\n\n')
exit(f'\t\t\t Duplicates exist, this has to be fixed, exiting at {line()}\n\n\n')
# Integrated from original: nan_alert()
def nan_alert(df_in, col=None):
''' nan stands for not-a-number, meaning null, check to see if any are present in the dataframe (df_in) '''
if not 'DataFrame' in str(type(df_in)):
print(f' nan_alert() expect a DataFrame, got {str(type(df_in))}, skipping')
return 0
count_nans = 0
if not len(df_in):
count_nans = 0
elif col is not None: # if column specified, otherwise entire df_in
df_partial = df_in.copy()[ ['VAERS_ID', col] ]
columns_w_nans = df_partial.columns[df_partial.isna().any()].tolist()
if col in columns_w_nans:
print(f'ERROR: {col} with Nans UNEXPECTED')
return 1
else:
columns_w_nans = df_in.columns[df_in.isna().any()].tolist()
if columns_w_nans:
print(f'\n\n\t nans in {columns_w_nans} line {inspect.stack()[1][2]}\n\n')
try:
df_with_nans = df_in[df_in.isna().any(axis=1)] # TODO: Truly just rows with nan? https://stackoverflow.com/questions/43424199/display-rows-with-one-or-more-nan-values-in-pandas-dataframe
count_nans = len(df_with_nans)
print(f' rows {count_nans}')
## TODO this, return df_in ... df_in.loc[df_in.VAERS_ID.isin(df_with_nans.VAERS_ID), 'trace' ] += f' . NaN'
except Exception as e:
print(df_in)
error(f"df_in[df_in.isna().any(axis=1)] {e}")
error(f'rows with NANS {inspect.stack()[1][2]}')
print(f'\n\n\n\n\t\t\t rows with NANS line {inspect.stack()[1][2]} \n {df_in} \n\n\n\n')
count_nans = 999999
return count_nans
# Integrated from original: lookup()
def lookup(previous_date, date_currently, vid_cols_affected):
''' A tool for adhoc on-the-fly debugging while at a breakpoint.
Show values for just the fields and VAERS_IDs for these compared files in dir_compared '''
print()
print(f'debug lookup() on vids affected')
files_populate_information()
df_changes = pd.DataFrame()
files_list = [ files['changes']['keyval'][previous_date], files['changes']['keyval'][date_currently] ]
vids_all_list = list(vid_cols_affected.keys())
cols_all = []
for k in vid_cols_affected:
for col in vid_cols_affected[k]:
if col not in cols_all:
cols_all.append(col)
for filename in files_list:
df_tmp = open_file_to_df(filename, doprint=0).fillna('') # [vids_all_list]
df_tmp = df_tmp.loc[ df_tmp.VAERS_ID.isin(vids_all_list) ]
df_tmp = df_tmp.copy()[['VAERS_ID'] + cols_all]
df_tmp['date'] = date_from_filename(filename)
df_changes = pd.concat([df_changes.reset_index(drop=True), df_tmp.reset_index(drop=True) ], ignore_index=True)
df_changes = fix_date_format(df_changes)
df_changes = move_column_forward(df_changes, 'date')
df_changes = df_changes.sort_values(by=['VAERS_ID', 'date'])
write_to_csv(df_changes, 'lookup_changes.csv', open_file=1)
print()
# Integrated from original: do_replace()
def do_replace(d, col, tag, this, that):
''' Not essential, a utility for testing some prep for harvesting numbers.
Makes regular expression replacements to remove false positives. Pared way down from its original elsewhere. '''
d2 = d.copy() # ???
print( f' do_replace {col} {tag:>40} {this:>40} {that}' )
d2[col] = d2[col].str.replace(this, that, flags=re.IGNORECASE, regex=True)
return d2
# Integrated from original: symptoms_file_entries_append_to_symptom_text()
def symptoms_file_entries_append_to_symptom_text(df_symfile):
''' Append symptoms entries from SYMPTOMS files to SYMPTOM_TEXT in symptoms column
Files like 2023VAERSSYMPTOMS.csv '''
for col in list(df_symfile.copy().columns): # limit to only SYMPTOM1 thru SYMPTOM5
if 'VAERS_ID' in col:
continue
if 'SYMPTOM' not in col:
del df_symfile[col]
continue
if 'VERSION' in col:
del df_symfile[col]
del col
''' Combine the symptoms column values to one string in a column called 'symptom_entries'
Step 1: Five columns to just one '''
print(' Combining symptoms column items. Grouping with delimiters, single row per VAERS_ID ...')
cols_symfile = sorted(set(df_symfile.columns) - set(['VAERS_ID']))
print(' Appending each symptom in new column called symptom_entries')
df_symfile['symptom_entries'] = ['_|_'.join(x) for x in np.sort(df_symfile[cols_symfile])] # Works. Combining the 5 columns as 1, keeping their order.
''' Step 2: Multiple VAERS_ID rows to just 1 row each '''
df_symfile = df_symfile.reset_index(drop=True) # avoid sort here
df_symfile = df_symfile[['VAERS_ID', 'symptom_entries']]
df_symfile = df_symfile.astype(str).groupby('VAERS_ID').agg(list).map('_|_'.join).reset_index()
#df_symfile = df_symfile.groupby('VAERS_ID').agg(lambda grp: '_|_'.join(grp.unique())).reset_index() # do not use drop=True here, it drops VAERS_ID
print(' Cleaning multiple delimiters due to empty columns')
df_symfile['symptom_entries'] = df_symfile.symptom_entries.str.replace(r'\s*_\|_\s*(?:\s*_\|_\s*)+\s*', '_|_', False, regex=True) # multiples to single (where empty columns)
''' _|_Always at beginning and end_|_ ... like a box or cell '''
df_symfile['symptom_entries'] = df_symfile.symptom_entries.str.replace(r'^(?!_\|_)(.*)$', r'_|_\1', False, regex=True) # _|_ at start of line always
df_symfile['symptom_entries'] = df_symfile.symptom_entries.str.replace(r'^(.*)(?!_\|_)$', r'\1_|_', False, regex=True) # _|_ at end of line always
df_long_cells = df_symfile.loc[ df_symfile.symptom_entries.str.len() >= 32720 ] # "Both . xlsx and . csv files have a limit of 32,767 characters per cell"
if len(df_long_cells):
print(f'{len(df_long_cells):>10} over 32720 in length to be truncated')
df_long_cells['symptom_entries'] = df_long_cells.symptom_entries.str.replace(r'^(.{32720,}).*', r'\1 \[truncated, Excel cell size limit 32,767\]', regex=True)
df_symfile = copy_column_from_1_to_2_per_vids('symptom_entries', df_long_cells, df_symfile, df_long_cells.VAERS_ID.to_list()) # TODO: This was 'symptoms', was a bug, right?
del df_long_cells
return df_symfile
# Integrated from original: make_numeric()
def make_numeric(df_in, col):
# There's been a lot of struggling with types and in the hands of garyha (me) in 2025 restart, still not all solved.
if not len(df_in): return df_in
df_in = df_in.copy()
df_in[col] = df_in[col].fillna('')
df_in[col] = df_in[col].astype(str)
try:
df_in[col] = pd.to_numeric(df_in[col])
except Exception as e:
print(f'pd.to_numeric Exception {e}')
# Desperation, number columns usually should be empty when 0, setting to 0.0 during run, empty them later
df_in.loc[ (df_in[ col ].isna()), col ] = 0.0
df_in[ col ] = df_in[ col ].astype('float64').round(4)
if 'nan' in df_in[col].to_list():
print(f'is_nan in make_numeric()')
return df_in
# Integrated from original: copy_column_from_1_to_2_per_vids()
def copy_column_from_1_to_2_per_vids(column, df1, df2, vids): # vids are VAERS_IDs
for col in df1.columns:
if col not in df2.columns:
df2[col] = '' # Avoid nans adding column empty string if didn't already exist
print(f' {col} made empty string in df2')
df2_sequestered_not_vids = df2.loc[ ~df2.VAERS_ID.isin(vids) ]
df1 = df1.loc[ df1.VAERS_ID.isin(vids) ]
df2 = df2.loc[ df2.VAERS_ID.isin(vids) ]
list_vids_both_only_to_copy = sorted( set(df1.VAERS_ID.to_list()) & set(df2.VAERS_ID.to_list()) )
df2 = df2.copy()
df2.loc[df2.VAERS_ID.isin( list_vids_both_only_to_copy ), column] = df2['VAERS_ID'].map(
pd.Series(df1[column].values, index=df1.VAERS_ID).to_dict()
)
if len(df2_sequestered_not_vids):
# Add back any that were in 2 but not in vids
df2 = pd.concat([
df2.reset_index(drop=True),
df2_sequestered_not_vids.reset_index(drop=True),
], ignore_index=True)
return df2
# To be untouched
df1_sequestered_not_vids = df1.loc[ ~df1.VAERS_ID.isin(vids) ]
df2_sequestered_not_vids = df2.loc[ ~df2.VAERS_ID.isin(vids) ]
df1 = df1.loc[ df1.VAERS_ID.isin(vids) ]
df2 = df2.loc[ df2.VAERS_ID.isin(vids) ]
# df1 and df2 are now only those that both have `vids` but this has become a mess, TODO
# Only apply to common rows
df1_not_in_df2 = df1.loc[ ~df1.VAERS_ID.isin(df2.VAERS_ID) ]
df2_not_in_df1 = df2.loc[ ~df2.VAERS_ID.isin(df1.VAERS_ID) ]
list_vids_both_only_to_copy = sorted( set(df1.VAERS_ID.to_list()) & set(df2.VAERS_ID.to_list()) )
df1_common_subset = df1.loc[ df1.VAERS_ID.isin(list_vids_both_only_to_copy) ]
df2_common_subset = df2.loc[ df2.VAERS_ID.isin(list_vids_both_only_to_copy) ]
# Make the changes
df2_common_subset = df2_common_subset.copy()
print(f'copy_column {column} on {len(list_vids_both_only_to_copy)} common vids. {len(df1_not_in_df2)} extra in df1, {len(df2_not_in_df1)} extra in df2')
df2_common_subset.loc[df2_common_subset.VAERS_ID.isin( list_vids_both_only_to_copy ), column] = df2_common_subset['VAERS_ID'].map(
pd.Series(df1_common_subset[column].values, index=df1_common_subset.VAERS_ID).to_dict()
)
if len(df1_not_in_df2) or len(df2_not_in_df1):
# Add back any that were in 2 but not in 1
df2 = pd.concat([
df1_not_in_df2.reset_index(drop=True),
df2_common_subset.reset_index(drop=True),
df2_not_in_df1.reset_index(drop=True),
df2_sequestered_not_vids.reset_index(drop=True),
], ignore_index=True)
return df2
# Integrated from original: move_rows()
def move_rows(df_subset, df_move_from, df_move_to):
''' Move rows in df_subset out of df_move_from into df_move_to
Return the new df_move_from and df_move_to '''
if not len(df_subset) or not len(df_move_from):
return df_move_from, df_move_to
df_move_to = df_move_to.copy()
'''
<string>:1: FutureWarning: The behavior of DataFrame concatenation with empty or all-NA entries is deprecated.
In a future version, this will no longer exclude empty or all-NA columns when determining the result dtypes.
To retain the old behavior, exclude the relevant entries before the concat operation.
'''
if not len(df_move_to): # avoiding that warning above
df_move_to = df_subset.copy()
else:
df_move_to = pd.concat([df_move_to.reset_index(drop=True), df_subset.reset_index(drop=True) ], ignore_index=True)
df_move_from = df_move_from.loc[ ~df_move_from.VAERS_ID.isin( df_move_to.VAERS_ID ) ] # everything from before, not now in done
return df_move_from, df_move_to
# Integrated from original: move_column_forward()
def move_column_forward(df_in, column):
''' Reorder columns moving column to second '''
if column not in df_in: return df_in
columns_pre = list(df_in.columns)
if columns_pre[1] == column: return df_in # already in that spot
if 'VAERS_ID' not in df_in.columns:
col_order = [column] # start
for col in df_in.columns:
if col == column: continue
col_order.append(col)
return df_in[col_order] #df_in.reindex(col_order, axis=1)
col_order = ['VAERS_ID', column]
for c in columns_pre:
if c not in col_order:
col_order.append(c)
return df_in.reindex(col_order, axis=1)
# Integrated from original: check_dupe_vaers_id()
def check_dupe_vaers_id(df):
if df is None or (not len(df)):
return 0
if not 'VAERS_ID' in df.columns:
return 0
dupes_list = [ x for x,count in Counter(df.VAERS_ID).items() if count > 1 ]
if dupes_list:
print(f'\n\nline {inspect.stack()[ 1 ][ 2 ]} WARNING: {len(dupes_list)} D U P L I C A T E VAERS_IDs: {subrange(dupes_list, 6)}\n\n')
return 1
return 0
# Integrated from original: prv_new_error_check()
def prv_new_error_check(df_prv, df_new):
''' Ensuring the VAERS_IDs are identical in each '''
vids_in_prv = df_prv.VAERS_ID.to_list()
vids_in_new = df_new.VAERS_ID.to_list()
if sorted(set(vids_in_prv)) != sorted(set(vids_in_new)):
print() ; print(f' ERROR: Expected VAERS_ID in prv and new to match exactly')
print(f' len prv: {len(vids_in_prv)}')
print(f' len new: {len(vids_in_new)}')
print(f' Difference: {len(vids_in_new) - len(vids_in_prv)}')
vids_in_prv_not_in_new = df_prv.loc[ ~df_prv.VAERS_ID.isin(df_new.VAERS_ID) ].VAERS_ID.to_list()
vids_in_new_not_in_prv = df_new.loc[ ~df_new.VAERS_ID.isin(df_prv.VAERS_ID) ].VAERS_ID.to_list()
print(f' In new not in prv: {vids_in_new_not_in_prv}')
print(f' In prv not in new: {vids_in_prv_not_in_new}')
print()
return 1
elif sorted(vids_in_prv) != sorted(vids_in_new): # as lists
print() ; print(f' ERROR: VAERS_ID in prv are the same sets but there are multiple VAERS_ID in one')
print(f' len prv: {len(vids_in_prv)}')
print(f' len new: {len(vids_in_new)}')
print(f' Difference: {len(vids_in_new) - len(vids_in_prv)}')
list_multiples_prv = [k[0] for k,v in df_prv[ ['VAERS_ID'] ].value_counts().to_dict().items() if v > 1]
if list_multiples_prv:
print(f' VAERS_IDs: {list_multiples_prv}')
print(f" df_prv.loc[ df_prv.VAERS_ID.isin({list_multiples_prv}) ][['VAERS_ID', 'VAX_LOT', 'VAX_MANU', 'VAX_TYPE', 'VAX_NAME']]")
df_multiples_prv = df_prv.loc[df_prv.VAERS_ID.isin(list_multiples_prv)][['VAERS_ID', 'VAX_LOT', 'VAX_MANU', 'VAX_TYPE', 'VAX_NAME']]
print(f'{df_multiples_prv}')
list_multiples_new = [k[0] for k,v in df_new[ ['VAERS_ID'] ].value_counts().to_dict().items() if v > 1]
if list_multiples_new:
print(f' VAERS_IDs: {list_multiples_new}')
print(f"df_new.loc[ df_new.VAERS_ID.isin({list_multiples_new}) ][['VAERS_ID', 'VAX_LOT', 'VAX_MANU', 'VAX_TYPE', 'VAX_NAME']]")
df_multiples_new = df_new.loc[ df_new.VAERS_ID.isin(list_multiples_new) ][['VAERS_ID', 'VAX_LOT', 'VAX_MANU', 'VAX_TYPE', 'VAX_NAME']]
print(f'{df_multiples_new}')
exit(' Cannot continue with this discrepancy')
# Integrated from original: debug_breakpoint()
def debug_breakpoint( date, col, vids, debug_dates, debug_cols, debug_vids ):
debug_pause = 0
if date in debug_dates or not debug_dates:
debug_pause += 1
if col in debug_cols or not debug_cols:
debug_pause += 1
if debug_vids:
vid_pause=0
for dv in debug_vids:
if dv in vids:
vid_pause = 1
break
if vid_pause:
debug_pause += 1
if debug_pause >= 3:
return 1 # a line for setting a breakpoint if conditions match
return 0
# Integrated from original: diff_context()
def diff_context( prv, new, col, vid, date_currently ):
''' Inputs: String column values like in SYMPTOM_TEXT. Code reviewer: Is there a better way below? Surely. '''
delim = ''
if len(prv) > 200:
delim_count, delim = get_prevalent_delimiters(prv)
a = diff_simplify( prv )
b = diff_simplify( new )
''' Ignore punctuation and upper/lower case. [^a-zA-Z0-9|] clears everything not alphanumeric or pipe symbol (since VAX delim matters there) '''
if re.sub(r'[^a-zA-Z0-9|]', '', a.lower()) == re.sub(r'[^a-zA-Z0-9|]', '', b.lower()):
return( '', '' )
if (delim and delim_count >= 5) or col == 'symptom_entries':
if col == 'symptom_entries': # these are lightning fast
delim = '_|_'
list_prv = a.split(delim)
list_new = b.split(delim)
only_prv = [x for x in list_prv if x not in list_new] # remove common
only_new = [x for x in list_new if x not in list_prv]
a = delim.join(only_prv)
b = delim.join(only_new)
''' Neutralizing punctuation changes needed here too because like 1334938 comma to pipe ... Hallucination| auditory ... wacky stuff by them '''
if re.sub(r'[^a-zA-Z0-9|]', '', a.lower()) == re.sub(r'[^a-zA-Z0-9|]', '', b.lower()):
return( '', '' ) # Ignore punctuation and upper/lower case
if delim == '_|_':
if sorted(only_prv) == sorted(only_new):
return ('', '') # Ignore reversed order
if a: a = delim + a + delim # endings
if b: b = delim + b + delim
return( a, b )
context = 6 # way low for cases like 1589954 to rid the word 'appropriate' by itself at len(largest_common_string) > context
count_while = 0
continue_while = 1
#length_max = 50 deprecated
while count_while <= 9 and continue_while: # Whiddling strings down, removing common substrings
count_while += 1 # Also consider if a in b: re.sub(a, '', b) ; a = '' and reverse for shorter no context
if not a or not b:
return( a, b )
if a.lower() == b.lower():
return( '', '' )
''' Important: If changes are the same with words just in a different order, skipping those.
Why? There are so many they would drown out changes. Applies for example to dose order changes, they are legion. '''
if delim:
if delim == '|' and sorted(set(a.split('|'))) == sorted(set(b.split('|'))): # vax fields, their delimiter is the pipe symbol, also removing duplicates
return( '', '' )
elif delim != '. ' and sorted(set(a.split(' '))) == sorted(set(b.split(' '))): # ignore reversed order and repeat words
return( '', '' )
''' To remove punctuation crudely: from string import punctuation ; ' '.join(filter(None, (word.strip(punctuation) for word in a.split()))) https://stackoverflow.com/a/15740656/962391
Ok, I give up. To avoid boatloads of problems, like commas replaced with pipe symbols in SYMPTOM_TEXT, going to compare/return just bare words for long strings
Avoid affecting things like 48.0
`punctuation` is just a string ... '!"#$%&\'()*+,-./:;<=>?@[\\]^_`{|}~'
punctuations = '!"#$%&()*+,-./:;<=>?@[\\]^_`{|}~' ... removing the single quote, note the 's', plural, variable name. '''
if len(a) > context and len(b) > context:
a = ' '.join(filter(None, (word.strip(''.join(punctuation)) for word in a.split()))) # consider not stripping ' .. ' instead but dicey
b = ' '.join(filter(None, (word.strip(''.join(punctuation)) for word in b.split())))
largest_string = longest_word_string(a, b)
if largest_string and (len(largest_string) > context): # and ((len(a) > length_max) or (len(b) > length_max)):
a = re.sub(re.escape(largest_string), ' .. ', a, flags=re.IGNORECASE)
b = re.sub(re.escape(largest_string), ' .. ', b, flags=re.IGNORECASE)
if a.lower() == b.lower():
return ('', '')
a_list = [x.strip() for x in a.split()] ; b_list = [x.strip() for x in b.split()]
only_a = [x for x in a_list if x not in b_list]
only_b = [x for x in b_list if x not in a_list]
a_list = [] ; b_list = []
for x in only_a:
if x in a_list:
continue
a_list.append(x)
for x in only_b:
if x in b_list:
continue
b_list.append(x)
a = ' '.join(a_list) ; b = ' '.join(b_list)
if not a or not b:
return (a, b)
else:
continue_while = 0
a = re.sub(r'\s\s+', ' ', a) ; b = re.sub(r'\s\s+', ' ', b) # multiple spaces
a = a.strip() ; b = b.strip() # front and end spaces
# remove multiples like ' cms hcc .. cms hcc .. cms hcc .. cms hcc '
if ' .. ' in a or ' .. ' in b:
a_list = a.split('..') ; b_list = b.split('..') # not a complete job on 1371552 but good enough for now
a_list = [x.strip() for x in a_list] ; b_list = [x.strip() for x in b_list]
# drop '' and ' '
a_list = [x for x in a_list if re.search(r'\w', x)] ; b_list = [x for x in b_list if re.search(r'\w', x)]
if len(a_list) != len(set(a_list)) or len(b_list) != len(set(b_list)): # if multiples
only_a = [x for x in a_list if x not in b_list]
only_b = [x for x in b_list if x not in a_list]
a_list = [] ; b_list = []
for x in only_a:
if x in a_list:
continue
a_list.append(x)
for x in only_b:
if x in b_list:
continue
b_list.append(x)
a = ' .. '.join(a_list) ; b = ' .. '.join(b_list)
a = re.sub(r'\s\s+', ' ', a) ; b = re.sub(r'\s\s+', ' ', b) # necessary anymore after additions above?
if re.sub(r'[^a-zA-Z0-9|]', '', a.lower()) == re.sub(r'[^a-zA-Z0-9|]', '', b.lower()): # again
return( '', '' ) # Ignore punctuation and upper/lower case
return ( a, b )
# Integrated from original: diff_simplify()
def diff_simplify( x ):
''' Remove trivial stuff from string, etc '''
x = re.sub(r' `\^` ' , '' , x) # the repeat sentence replacements that were entered
x = re.sub(r'[^\x00-\x7F]+', ' ' , x) # unicode
x = re.sub(r'\s\s+' , ' ' , x) # multiple spaces with single to avoid trivial
x = re.sub(r"''" , "'" , x) # doubled can mess things up like ... I''m ... in 1455992
x = x.strip() # front and end spaces
return x
# Integrated from original: get_prevalent_delimiters()
def get_prevalent_delimiters(prv_str):
''' '''
max_delim = 0
delim = ''
for d in [ '. ', '|', '; ', ' - ' ]:
if d == '|' and '_|_' in prv_str:
continue # clumsy but these get crossed and '_|_' handled elsewhere
count = prv_str.count(d)
if count > max_delim:
max_delim = count
if max_delim >= 5:
delim = d
return( max_delim, delim )
# Integrated from original: longest_word_string()
def longest_word_string(a, b):
''' https://stackoverflow.com/a/42882629/962391 '''
answer = []
list_a = a.split()
list_b = b.split()
len1, len2 = len(list_a), len(list_b)
for i in range(len1):
for j in range(len2):
lcs_temp = 0
match = []
while ((i+lcs_temp < len1) and (j+lcs_temp < len2) and list_a[i+lcs_temp].lower() == list_b[j+lcs_temp].lower()):
match.append( list_b[j+lcs_temp].lower() )
lcs_temp += 1
if len(match) > len(answer):
answer = match
answer = ' '.join(answer)
answer = answer.strip()
return answer
# Integrated from original: find_all_context()
def find_all_context(search_string, df_in, df_in_column, df_out_column):
search_string = search_string .lower()
df_in[df_in_column] = df_in[df_in_column].str.lower()
df_found = df_in.loc[ df_in[df_in_column].str.contains(search_string, na=False) ]
if len(df_found):
df_found = df_found.copy()
str_context = r'\b(.{0,20}' + search_string + r'.{0,20})\b'
df_found[df_out_column] = df_found[df_in_column].str.findall(str_context).str.join(' ~~ ')
return df_found
else:
return pd.DataFrame() # empty for len 0
# Integrated from original: do_autodownload()
def do_autodownload():
''' https://vaers.hhs.gov/data/datasets.html '''
if not autodownload:
return
sys.path.insert(0, './vaers_downloader')
# enable when the time comes # from VAERSFileDownloader import updateVAERSFiles
updateVAERSFiles(
needsUpdate = True,
years = [2020],
workingDirectory = os.getcwd()
)
# Integrated from original: symptoms_dedupe_repeat_sentences()
def symptoms_dedupe_repeat_sentences(_data):
''' Remove repeat sentences '''
print(' Repeat sentence removal in SYMPTOM_TEXT (takes time)') # Will anyone be unhappy that SYMPTOM_TEXT are no longer pristine but deduped?
_data = _data.copy()
## tmp replacing old ...^... with `^`
#syms_with_old_placeholder = _data.loc[_data.SYMPTOM_TEXT.str.contains(r'\.\.\.\^\.\.\.', na=False)]
#if len(syms_with_old_placeholder):
# print() ; print(f'{len(syms_with_old_placeholder):>10} old placeholders in symptoms_dedupe_repeat_sentences, replacing with `^`') ; print()
if len(_data):
#if len(syms_with_old_placeholder):
# syms_with_old_placeholder['SYMPTOM_TEXT'] = syms_with_old_placeholder.SYMPTOM_TEXT.str.replace(r'\.\.\.\^\.\.\.', ' `^` ')
# _data = copy_column_from_1_to_2_per_vids('SYMPTOM_TEXT', syms_with_old_placeholder, _data, syms_with_old_placeholder.VAERS_ID.to_list())
_data['SYMPTOM_TEXT'] = _data.apply(lambda row: symptoms_dedupe_repeat_sentences_each( row[['VAERS_ID', 'SYMPTOM_TEXT']] ), axis=1)
print()
to_print = f"{stats['dedupe_count']:>10} SYMPTOM_TEXT field repeat sentences deduped in "
to_print += f"{stats['dedupe_reports']} reports, max difference {stats['dedupe_max_bytes']} bytes in VAERS_ID {stats['dedupe_max_vid']}"
print(to_print) ; print()
return _data
# Integrated from original: symptoms_dedupe_repeat_sentences_each()
def symptoms_dedupe_repeat_sentences_each(series_vid_sym):
''' Remove repeat sentences in VAERS SYMPTOM_TEXT fields
Prints each time a larger change occurs.
Originated at https://stackoverflow.com/a/40353780/962391 '''
# Use delim_count, delim = get_prevalent_delimiters(prv)
# In 2025 ... :1818: FutureWarning: Series.__getitem__ treating keys as positions is deprecated.
# In a future version, integer keys will always be treated as labels (consistent with DataFrame behavior).
# To access a value by position, use `ser.iloc[pos]`
vid = int(series_vid_sym.iloc[0]) # 2025 force int, is this a mistake? TODO.
content = str(series_vid_sym.iloc[1]) # sometimes not string
''' Some of the input files filled in by various people (at CDC) evidently might have gone through some processing in Excel by them.
As a result, fields have double quotes injected, they have to be removed or big trouble, takes time but required. '''
content = re.sub(r'^"', '', content) # not certain about these
content = re.sub(r'"$', '', content)
delim = get_prevalent_delimiters(content)[1]
if not delim:
return content
list_in = re.split(re.escape(delim), content)
unique_set = set(list_in)
dupe_count = len(list_in) - len(unique_set)
if not dupe_count:
return content # no dupes
count_of_replacements = 0
total_bytes = 0
list_out = []
for line in list_in:
#if '`^`' in line: # previously processed by mistake?
# continue
if line in list_out and (len(line) > 40): # only dedupe longish lines
#try:
# print(f' {line}')
#except Exception as e:
# pass # UnicodeEncodeError: 'charmap' codec can't encode character '\ufffd' in position 4: character maps to <undefined>
count_of_replacements += 1
total_bytes += len(line)
if vid not in writeups_deduped:
stats['dedupe_bytes'] += len(line)
line = '`^`'
list_out.append(line)
if total_bytes > stats['dedupe_max_bytes']:
stats['dedupe_max_bytes'] = total_bytes
stats['dedupe_max_vid' ] = vid
content = delim.join(list_out)
# stats
if vid not in writeups_deduped: # count each report only once, first time seen
stats['dedupe_reports'] += 1 # count of reports that ever had any repeat sentences
writeups_deduped[vid] = 1 # for unique vid tracking
#count_of_replacements = content.count('`^`')
if count_of_replacements:
stats['dedupe_count'] += count_of_replacements
return content # modified
# - - - - - - - - - - - - - - -
# BELOW SEEMS TO HAVE FLAWS
# string_split_multiple_delims = re.split('[:;,\|]\s*', content) # doesn't preserve them
list_in = re.split(r'[^a-zA-Z0-9\'\s]', content) # split on everything except alphanumeric, space etc
list_in = [x for x in list_in if len(x) >= 40] # just the longer strings
dupes_list = [x for x, count in Counter(list_in).items() if count > 1]
if not dupes_list:
return content
# Doing dedupe counts only once per VAERS_ID
# Replace all but the first with `^` , low tech solution: https://stackoverflow.com/a/53239962/962391
total_bytes = 0
for dup in dupes_list:
content = content.replace(re.escape(dup), ' `^` ').replace(' `^` ', dup, 1)
total_bytes += len(dup)
if vid not in writeups_deduped:
stats['dedupe_bytes'] += len(dup)
if total_bytes > stats['dedupe_max_bytes']:
stats['dedupe_max_bytes'] = total_bytes
stats['dedupe_max_vid' ] = vid
# stats
if vid not in writeups_deduped: # count each report only once, first time seen
stats['dedupe_reports'] += 1 # count of reports that ever had any repeat sentences
writeups_deduped[vid] = 1 # for unique vid tracking
count_of_replacements = content.count(' `^` ')
if count_of_replacements:
stats['dedupe_count'] += count_of_replacements
return content # modified
# Integrated from original: get_next_date()
def get_next_date():
''' Next date has to be the next flattened needed to be done. Compare uses flattened files. '''
# TODO: This has changed, there's surely some dead code here now that would never be hit, to be removed. Also find ... cheap trick, this is part of it. Slopppy ...
dates_input = sorted( [x for x in files['input' ]['date'] if x not in files['done']]) # added 'done' in 2025
dates_changes = sorted(files['changes']['date'])
if not files['flattened']['keyval']: # first run
return dates_input[0]
# Comparing to inputs (to do)
if dates_changes:
high_changes_done = dates_changes[-1]
changes_next_candidates = [x for x in dates_input if x > high_changes_done]
if changes_next_candidates:
date_next = changes_next_candidates[0]
if date_ceiling and ( date_next > date_ceiling ):
exit(f'\n\n\n\n\t\t\t date_ceiling is set and was reached: date_next {date_next} > {date_ceiling} \n\n\n\n')
return date_next
else:
''' Save final output as two, split in half, due to Excel row limit of about 1.048 million per sheet. '''
# dead no? if len(df_flat_prv):
# dead no? print() ; print(' save_multi_csv(high_changes_done, df_flat_prv)') ; print()
# dead no? save_multi_csv(high_changes_done, df_flat_prv)
if date_ceiling:
exit(f'\n\n\n\n\t\t\t date_ceiling is {date_ceiling}, highest changes: {high_changes_done} \n\n\n\n')
else:
print()
exit(' No more input files to process') ; print()
else:
''' Confusions. Whole thing needs a lot of conditions applied.
If there are no changes files, can still be consolidated and flattened.
If no changes, just return the first as next? No, that's a loop.
'''
if files['flattened']['date']:
if dates_input[0] in files['flattened']['date']:
# Initiating, copy it
date_currently = dates_input[0]
print_date_banner(date_currently)
file_for_copy_original = dir_compared + '/' + date_currently + '_VAERS_FLATFILE.csv'
print(
f' Due to first drop, creating from flattened: {file_for_copy_original}')
print()
file_flattened = files['flattened']['keyval'][date_currently]
shutil.copy(file_flattened, file_for_copy_original) # copyfile() doesn't include permissions so it can result in problems
# The file has to be read for VAERS_IDs for ever_covid to be accurate
print() ; print(f' Reading original for ever_covid bookkeeping')
df = open_file_to_df(file_flattened, doprint=1)
# Initial row in stats file when only one compare/changes file
stats_initialize (date_currently) # per drop, then a totals row is calculated
do_never_ever( df.VAERS_ID.to_list(), date_currently, 'get_next_date on file_flattened' )
do_ever_covid( sorted(set(df.VAERS_ID.to_list())) )
stats_resolve(date_currently)
return dates_input[1] # NEXT ONE
return dates_input[0]
consolidated_not_done = sorted( set(files['input']['date']) - set(files['consolidated']['date']) )
if consolidated_not_done:
date_todo = sorted(consolidated_not_done)[0]
print(f' Consolidation to do: {date_todo}')
return date_todo
flattened_not_done = sorted( set(files['input']['date']) - set(files['flattened']['date']) )
if flattened_not_done:
date_todo = sorted(flattened_not_done)[0]
print(f' Flattening to do: {date_todo}')
return date_todo
latest_flattened_done = sorted(files['flattened']['date'])[-1] if files['flattened']['date'] else ''
dates_list = [x for x in dates_input if x > latest_flattened_done]
date_next = dates_list[0]
if len(dates_list) == 0: # No more to process
print()
exit(' No more files to process')
elif date_ceiling and ( date_next > date_ceiling ):
exit(f'\n\n\n\n\t\t\t date_ceiling is set and was reached: date_next {date_next} > {date_ceiling} \n\n\n\n')
else:
return date_next
# Integrated from original: more_to_do()
def more_to_do():
''' Return 1 or 0 on whether any more input files process '''
global elapsed_drop, df_flat_1
files_populate_information()
if not files['changes']['date']: # None done yet
return 1
if files['input']['date'] == files['flattened']['date'] == files['consolidated']['date'] == files['changes']['date']:
if files['changes']['date'][-1] >= files['input']['date'][-1]:
date_currently = files['changes']['date'][-1]
if df_flat_1 is not None and len(df_flat_1):
pass
#save_multi_csv(date_currently, df_flat_1) # more than one csv due to Excel row limit
else:
prv_date = files['changes']['date'][-1]
filename = files['changes']['keyval'][prv_date]
print(f'Patching for creation of split changes file _A and _B, no more to do case unusual, using {filename}')
df_flat_1 = open_file_to_df(filename, doprint=0)
print() ; print(' save_multi_csv(date_currently, df_flat_1)') ; print()
save_multi_csv(date_currently, df_flat_1) # more than one csv due to Excel row limit
print()
print(f" No more to do, last set {files['changes']['date'][-1]} >= {files['input']['date'][-1]} done")
return 0
elapsed_drop = _time.time() # new start marker for total time on each drop
return 1
# Integrated from original: print_date_banner()
def print_date_banner(this_drop_date):
print()
print('= = = = = = = = = = = = = = = = = = = = = = = = = = = = = = ')
print(f' Next date {this_drop_date}')
print('= = = = = = = = = = = = = = = = = = = = = = = = = = = = = = ')
print()
# Integrated from original: consolidate()
def consolidate(files_date_marker):
'''
= = = = = = = = = = = = = = = = = = = = = = = = = = = = = =
Consolidate -- All in one file but still multiple rows per VAERS_ID sometimes.
= = = = = = = = = = = = = = = = = = = = = = = = = = = = = = '''
global df_vax, df_data, df_syms_flat, ever_covid, ever_any, covid_earliest_vaers_id
print() ; print(f' Consolidation')
files_populate_information()
''' Consolidate only if the file doesn't already exist '''
if files_date_marker in files['consolidated']['date']:
if files_date_marker in files['consolidated']['keyval']: # already done, use it
print(f" Consolidation already done: {files['consolidated']['keyval'][files_date_marker]}")
df_vax = pd.DataFrame() # avoid picking up a previous consolidation when flattening
return
else:
print(f" ERROR: Expected {files_date_marker} in files['consolidated']['keyval']")
if files_date_marker in files['flattened']['date']:
print(f' Skipping consolidation because flattened for {files_date_marker} already exists')
return
''' Combine all data, vax and symptoms files as one
(if not already a file like 2020-12-25_VAERS_CONSOLIDATED.csv in which case just do the treament) '''
print(' Concatenating files, *VAERSDATA.csv, *VAERSVAX.csv, *VAERSSYMPTOMS.csv')
df_vax = files_concat( glob.glob(dir_working + '/' + '*VAERSVAX.csv' ) )
''' Remove all reports prior to the first covid jab. [That was the thinking but now in 2025 with all CDC drops (at last), widen to all vaccines?]
2020-12-14 902418 First report after public rollout is next day: https://www.medalerts.org/vaersdb/findfield.php?IDNUMBER=902418&WAYBACKHISTORY=ON
2020-10-02 896636 may have been from a trial: https://www.medalerts.org/vaersdb/findfield.php?IDNUMBER=896636&WAYBACKHISTORY=ON
VAERS ID: 896636
VAERS Form: 2
Age: 47.0
Sex: Female
Location: South Carolina
Vaccinated: 2020-09-28
Onset: 2020-10-02
Submitted: 0000-00-00
Entered: 2020-11-14
Vaccination / Manufacturer (1 vaccine) Lot / Dose Site / Route
COVID19: COVID19 (COVID19 (MODERNA)) / MODERNA - / UNK LA / SYR
2020-12-10 970043 Died same day, reported later: https://www.medalerts.org/vaersdb/findfield.php?IDNUMBER=970043&WAYBACKHISTORY=ON
'''
# Automatically detect earliest covid VAERS_ID. TODO: Do earlier in the code?
df_vax_all_covid = df_vax.loc[ df_vax[ 'VAX_TYPE' ].str.lower().str.contains('covid', na=False) ]
if len(df_vax_all_covid):
covid_earliest_vaers_id_now = df_vax_all_covid.VAERS_ID.astype(int).min()
if covid_earliest_vaers_id == 0: # no previous VAERS_ID value
covid_earliest_vaers_id = covid_earliest_vaers_id_now
print(f' Earliest covid VAERS_ID initiated at {covid_earliest_vaers_id}\n')
elif covid_earliest_vaers_id > covid_earliest_vaers_id_now:
# was previously set but is even lower now (altho published later)
print(f' Earliest covid VAERS_ID set EVEN LOWER now at {covid_earliest_vaers_id}, was {covid_earliest_vaers_id}\n')
covid_earliest_vaers_id = covid_earliest_vaers_id_now
else:
print(f' No covid found in {files_date_marker}\n')
# Cheap trick to avoid running the same input again, and this should be improved. Quite the logic puzzle. Trimming files_date_marker from inputs list.
files[ 'input' ][ 'date' ] = [x for x in files['input' ]['date'] if x != files_date_marker]
return
stats['lo_ever'] = covid_earliest_vaers_id # TODO: 2025, wrong? right?
df_syms = files_concat( glob.glob(dir_working + '/' + '*VAERSSYMPTOMS.csv') )
df_data_all = files_concat( glob.glob(dir_working + '/' + '*VAERSDATA.csv' ) )
if vids_limit: # in testing special case
df_data_all = df_data_all.loc[ df_data_all.VAERS_ID.isin(vids_limit) ]
df_vax = df_vax .loc[ df_vax .VAERS_ID.isin(vids_limit) ]
df_syms = df_syms .loc[ df_syms .VAERS_ID.isin(vids_limit) ]
len_before = len(df_data_all)
df_data_all = df_data_all.loc[ df_data_all.VAERS_ID >= covid_earliest_vaers_id ]
df_vax = df_vax .loc[ df_vax .VAERS_ID >= covid_earliest_vaers_id ]
df_syms = df_syms .loc[ df_syms .VAERS_ID >= covid_earliest_vaers_id ]
print() ; print(f'{len_before - len(df_data_all):>10} records removed prior to the first covid report (covid_earliest_vaers_id {covid_earliest_vaers_id})')
print(f'{len(df_data_all):>10} covid vax reports to work with (unique VAERS_IDs)') ; print()
vids_list = df_data_all.VAERS_ID.to_list()
lo_data_all = min(vids_list)
hi_data_all = max(vids_list)
diff_data_all = hi_data_all - lo_data_all
len_data_all = len(vids_list)
missing = diff_data_all - len_data_all
print(f'{missing:>10} missing (any/all vax never published in covid era) is implied by only {len_data_all} present in {diff_data_all} range with lo {lo_data_all} and hi {hi_data_all}')
print()
vids_new = [ x for x in vids_list if x not in ever_any ]
print(f'{len(vids_new):>10} being added to `ever_any` for this drop, any/all vax')
ever_any.update({x:1 for x in vids_list})
#ever_any = {**ever_any, **{x:1 for x in vids_list}}
#ever_any = {x:1 for x in set(ever_any.keys() + vids_list)}
# Grab all VAERS_IDs before filtering for covid, to be able to identify gaps properly
do_never_ever( vids_list, files_date_marker, 'consolidate on df_data_all' )
stats['drop_input_covid'] = len(df_data_all)
''' VAERS_IDs can have multiple records/doses/lots in each report but at least one covid19. '''
dfv = df_vax.copy()
dfv['doses'] = dfv.VAERS_ID.map(dfv.VAERS_ID.value_counts())
dfv_single_doses = dfv.loc[ dfv.doses.eq(1) ]
dfv_singles_w_covid = dfv_single_doses.loc[ dfv_single_doses.VAERS_ID.isin(dfv_single_doses.loc[ dfv_single_doses.VAX_TYPE.str.contains('COVID', na=False) ].VAERS_ID.to_list()) ].sort_values(by='VAERS_ID')
dfv_multpl_doses = dfv.loc[ dfv.doses.ge(2) ]
dfv_multiples_w_covid = dfv_multpl_doses.loc[ dfv_multpl_doses.VAERS_ID.isin(dfv_multpl_doses.loc[ dfv_multpl_doses.VAX_TYPE.str.contains('COVID', na=False) ].VAERS_ID.to_list()) ].sort_values(by='VAERS_ID')
del dfv
dfv_covid_type_both = pd.concat([dfv_singles_w_covid.reset_index(drop=True), dfv_multiples_w_covid.reset_index(drop=True) ], ignore_index=True)
dfv_covid_type_both = dfv_covid_type_both.drop_duplicates()
df_data_other_for_covid_search = df_data_all.loc[ ~df_data_all.VAERS_ID.isin(dfv_covid_type_both.VAERS_ID) ]
df_data_covid_other_found1 = df_data_other_for_covid_search.loc[ df_data_other_for_covid_search.SYMPTOM_TEXT.str.contains(r'Pfizer|Moderna|Janssen', re.IGNORECASE, na=False) ]
df_data_covid_other_found2 = df_data_covid_other_found1 .loc[ df_data_covid_other_found1 .SYMPTOM_TEXT.str.contains(r'Covid', re.IGNORECASE, na=False) ]
print() ; print(f'{len(df_data_covid_other_found2):>10} additional reports captured with Pfizer|Moderna|Janssen and Covid in SYMPTOM_TEXT although not officially type COVID') ; print()
vids_vax__covid_type_both = dfv_covid_type_both .VAERS_ID.to_list()
vids_data_covid_other_found = df_data_covid_other_found2.VAERS_ID.to_list()
vids_all_covid_list = sorted( set(vids_vax__covid_type_both + vids_data_covid_other_found) )
# Reducing to only covid reports
len_data_all_prev_any = len(df_data_all)
df_data = df_data_all.loc[ df_data_all.VAERS_ID.isin(vids_all_covid_list) ]
df_vax = df_vax .loc[ df_vax .VAERS_ID.isin(vids_all_covid_list) ]
df_syms = df_syms .loc[ df_syms .VAERS_ID.isin(vids_all_covid_list) ]
print(f'{ len_data_all_prev_any:>10} total any vax reports') ; print()
print(f'{len_data_all_prev_any - len(df_data):>10} non-covid vax reports excluded') ; print()
stats['drop_input_covid'] = len(df_data)
#do_ever_covid( vids_all_covid_list ) # Only covid reports, subset of ever_any which is all vax
len_before = len(df_data)
df_data = df_data.drop_duplicates(subset='VAERS_ID')
if len(df_data) - len_before:
print(f'{(len_before - len(df_data)):>10} duplicates dropped in df_data on VAERS_IDs')
print(f'{len(set(df_data.VAERS_ID.to_list())):>10} covid reports to work with') ; print()
len_before = len(df_data)
df_data = df_data.drop_duplicates(subset='VAERS_ID')
if len(df_data) - len_before:
print(f'{(len_before - len(df_data)):>10} duplicates dropped in df_data on VAERS_IDs, expected none')
''' Shorten some fields in VAX (the change to title case also indicates I've touched it) '''
print(' Shortening some field values in VAX_NAME, VAX_MANU') ; print()
'''
VAX_TYPE
COVID19
COVID19-2
FLU4
VAX_NAME Shorter
COVID19 (COVID19 (MODERNA)) C19 Moderna
COVID19 (COVID19 (MODERNA BIVALENT)) C19 Moderna BIVALENT
COVID19 (COVID19 (PFIZER-BIONTECH)) C19 Pfizer-BionT
COVID19 (COVID19 (PFIZER-BIONTECH BIVALENT)) C19 Pfizer-BionT BIVALENT
COVID19 (COVID19 (JANSSEN))
COVID19 (COVID19 (UNKNOWN))
COVID19 (COVID19 (NOVAVAX))
INFLUENZA (SEASONAL) (FLULAVAL QUADRIVALENT)
Transitional with parens removed first step
COVID19 COVID19 MODERNA
COVID19 COVID19 MODERNA BIVALENT
COVID19 COVID19 PFIZER-BIONTECH
COVID19 COVID19 PFIZER-BIONTECH BIVALENT
COVID19 COVID19 JANSSEN
INFLUENZA SEASONAL FLULAVAL QUADRIVALENT '''
df_vax = df_vax.copy()
df_vax['VAX_NAME'] = df_vax.VAX_NAME.str.replace(r'[\)\(]' , r'' , False, regex=True)
# COVID19 COVID19 ... and/or COVID19-2 COVID19-2 ... except those so far don't show up in VAX_NAME, just in case
df_vax['VAX_NAME'] = df_vax.VAX_NAME.str.replace(r'(?:COVID(\S+)\s)+' , r'C\1 ' , False, regex=True)
#df_vax['VAX_NAME'] = df_vax.VAX_NAME.str.replace(r'(?:COVID(\d+)(\-\d+)*\s)+' , r'C\1 \2' , False, regex=True)
df_vax['VAX_NAME'] = df_vax.VAX_NAME.str.replace(r'PFIZER.BION.*' , r'Pfizer-BionT' , False, regex=True)
df_vax['VAX_NAME'] = df_vax.VAX_NAME.str.replace(r'MODERNA' , r'Moderna' , False, regex=True)
df_vax['VAX_NAME'] = df_vax.VAX_NAME.str.replace(r'JANSSEN' , r'Janssen' , False, regex=True)
df_vax['VAX_NAME'] = df_vax.VAX_NAME.str.replace(r'INFLUENZA' , r'Flu' , False, regex=True)
df_vax['VAX_NAME'] = df_vax.VAX_NAME.str.replace(r'VACCINE NOT SPECIFIED' , r'Not Specified' , False, regex=True)
df_vax['VAX_MANU'] = df_vax.VAX_MANU.str.replace(r'UNKNOWN MANUFACTURER' , r'Unknown' , False, regex=True)
df_vax['VAX_MANU'] = df_vax.VAX_MANU.str.replace(r'PFIZER.BION.*' , r'Pfizer-BionT' , False, regex=True)
df_vax['VAX_MANU'] = df_vax.VAX_MANU.str.replace(r'MODERNA' , r'Moderna' , False, regex=True)
df_vax['VAX_MANU'] = df_vax.VAX_MANU.str.replace(r'JANSSEN' , r'Janssen' , False, regex=True)
df_vax['VAX_MANU'] = df_vax.VAX_MANU.str.replace(r'PF.*WYETH' , r'Pfizer-Wyeth' , False, regex=True)
df_vax['VAX_MANU'] = df_vax.VAX_MANU.str.replace(r'NOVARTIS.*' , r'Novartis' , False, regex=True)
df_vax['VAX_MANU'] = df_vax.VAX_MANU.str.replace(r'SANOFI.*' , r'Sanofi' , False, regex=True)
df_vax['VAX_MANU'] = df_vax.VAX_MANU.str.replace(r'MERCK.*' , r'Merck' , False, regex=True)
df_vax['VAX_MANU'] = df_vax.VAX_MANU.str.replace(r'PROTEIN.*' , r'Protein' , False, regex=True)
df_vax['VAX_MANU'] = df_vax.VAX_MANU.str.replace(r'GLAXO.*' , r'Glaxo' , False, regex=True)
df_vax['VAX_MANU'] = df_vax.VAX_MANU.str.replace(r'SEQIRUS.*' , r'Seqirus' , False, regex=True)
df_vax['VAX_MANU'] = df_vax.VAX_MANU.str.replace(r'BERNA.*' , r'Berna' , False, regex=True)
# To avoid jumbling in the case of tiny differences between drops, apparently this is necessary why?
df_vax = df_vax.sort_values(by=['VAERS_ID', 'VAX_LOT', 'VAX_SITE', 'VAX_DOSE_SERIES', 'VAX_TYPE', 'VAX_MANU', 'VAX_ROUTE', 'VAX_NAME'])
print(' Merging DATA into VAX')
df_data_vax = pd.merge(df_vax.astype(str), df_data.astype(str), on='VAERS_ID', how='left')
if nan_alert(df_data_vax):
pause=1 # for a breakpoint sometimes
df_data_vax = df_data_vax.fillna('')
print(f'{len(df_data_vax):>10} rows in df_data_vax (can be more than one row per VAERS_ID)')
'''
= = = = = = = = = = = = = = = = = = = = = = = = = = = = = =
symptom_entries
= = = = = = = = = = = = = = = = = = = = = = = = = = = = = =
'''
''' Aggregate symptoms into new column called symptom_entries, and merge for all symptoms covered as a single string '''
print(' Aggregating symptoms into symptom_entries string, new column')
df_syms_flat = symptoms_file_entries_append_to_symptom_text(df_syms)
'''
= = = = = = = = = = = = = = = = = = = = = = = = = = = = = =
Consolidation
Save result into one file like ... 2020-12-25_VAERS_CONSOLIDATED.csv
More than one row per VAERS_ID for the various doses/lots, thus duplicates to symptom_entries etc.
= = = = = = = = = = = = = = = = = = = = = = = = = = = = = =
'''
''' symptom_entries to consolidated '''
print(' Merging symptom_entries into df_data_vax')
df_data_vax_syms_consolidated = pd.merge(df_data_vax.astype(str), df_syms_flat.astype(str)[['VAERS_ID', 'symptom_entries']], on='VAERS_ID', how='left')
#nan_alert(df_data_vax_syms_consolidated) # nan in symptom_entries in 1106891 on 03/10/2021 as the symptoms file has no entry line for it even tho Current Illness: Atrial Fibrillation
df_data_vax_syms_consolidated = df_data_vax_syms_consolidated.fillna('')
print(f'{len(df_data_vax_syms_consolidated):>10} rows in df_data_vax_syms_consolidated') # has dupe VAERS_IDs as expected, more than one dose per report
filename_consolidated = dir_consolidated + '/' + files_date_marker + '_VAERS_CONSOLIDATED.csv'
print(f' Saving result into one file w/ multiple rows per VAERS_ID: {filename_consolidated}')
write_to_csv(df_data_vax_syms_consolidated, filename_consolidated)
print()
print(f' Consolidation of {files_date_marker} done')
return
# Integrated from original: flatten()
def flatten(files_date_marker):
'''
= = = = = = = = = = = = = = = = = = = = = = = = = = = = = =
Flattening -- Only one row per VAERS_ID
= = = = = = = = = = = = = = = = = = = = = = = = = = = = = = '''
global df_vax, df_data, df_syms_flat, df_flat_2
if not covid_earliest_vaers_id:
files['done'].append(files_date_marker)
return
print()
print(f' Flattening')
files_populate_information()
''' Flatten only if the file doesn't already exist '''
if files_date_marker in files['flattened']['date']:
if files_date_marker in files['flattened']['keyval']: # already done, use it
print(f" Flattening already done: {files['flattened']['keyval'][files_date_marker]}") ; print()
return
else:
print(f" ERROR: Expected {files_date_marker} in files['flattened']['keyval']")
pull_vax_records = 0
file_consolidated = ''
if not len(df_vax): # already exists if consolidate just created it
# vax file records from consolidate file
if files_date_marker in files['consolidated']['keyval']:
file_consolidated = files['consolidated']['keyval'][files_date_marker]
print(f" Pulling vax records from previously consolidated file {file_consolidated}")
pull_vax_records = 1
else:
print(f" ERROR: Expected {files_date_marker} in files['consolidated']['keyval']")
if pull_vax_records:
df_consolidated = open_file_to_df(file_consolidated, doprint=1).fillna('')
columns_vax = ['VAERS_ID', 'VAX_TYPE', 'VAX_MANU', 'VAX_LOT', 'VAX_DOSE_SERIES', 'VAX_ROUTE', 'VAX_SITE', 'VAX_NAME'] # Note VAX_DATE is in data, not vax file
columns_consolidated = df_consolidated.columns
df_vax = df_consolidated[columns_vax].astype(str)
columns_data = ['VAERS_ID'] + list( set(columns_consolidated) - set(columns_vax) )
df_data = df_consolidated[columns_data].astype(str)
del df_data['symptom_entries'] # added back in later
df_syms_flat = df_consolidated.astype(str)[['VAERS_ID', 'symptom_entries']]
del file_consolidated, pull_vax_records, columns_vax, columns_data
if not stats['drop_input_covid']:
stats['drop_input_covid'] = len(df_data)
print(f"=== Added count_input {stats['drop_input_covid']} in def flattening")
''' Aggregate VAX for single VAERS_ID '''
print(' Aggregate/flatten VAX items. Grouping by VAERS_ID')
df_vax = df_vax.reset_index(drop=True)
df_vax_flat = df_vax.groupby('VAERS_ID').agg(list).map('|'.join).reset_index()
del df_vax # done with this global
len_before = len(df_vax_flat)
df_vax_flat = df_vax_flat.drop_duplicates(subset=df_vax_flat.columns, keep='first')
if len(df_vax_flat) - len_before:
print(f'\n\n\n\n\t\t\t\t {(len_before - len(df_vax_flat)):>10} duplicates dropped in df_vax_flat, THERE SHOULD BE NONE\n\n\n\n')
check_dupe_vaers_id(df_vax_flat) ; print()
''' Combine DATA into VAX (has multiple records for each VAERS_ID, when more than one dose is listed) '''
print(' Merging DATA into VAX flattened')
df_data_vax_flat = pd.merge(df_vax_flat.astype(str), df_data.astype(str), on='VAERS_ID', how='left')
if nan_alert(df_data_vax_flat):
pause=1
df_data_vax_flat = df_data_vax_flat.fillna('')
check_dupe_vaers_id(df_vax_flat)
len_before = len(df_data_vax_flat)
df_data_vax_flat = df_data_vax_flat.drop_duplicates(subset=df_data_vax_flat.columns, keep='first')
if len(df_data_vax_flat) - len_before:
print(f'{(len_before - len(df_data_vax_flat)):>10} duplicates dropped in df_data_vax_flat')
print(f'{len(df_data_vax_flat):>10} rows in df_data_vax_flat') ; print()
''' symptom_entries to flattened '''
print(' Merging symptom_entries into df_data_vax_syms_flat')
df_data_vax_syms_flat = pd.merge(df_data_vax_flat.astype(str), df_syms_flat.astype(str)[['VAERS_ID', 'symptom_entries']], on='VAERS_ID', how='left')
df_data_vax_syms_flat = df_data_vax_syms_flat.fillna('') # there are nans like 1106891 with no record for it in ...symptoms.csv
del df_syms_flat
len_before = len(df_data_vax_syms_flat) # TODO: Understand the reason for the dupes
df_data_vax_syms_flat = df_data_vax_syms_flat.drop_duplicates(subset=df_data_vax_syms_flat.columns, keep='first')
if len(df_data_vax_syms_flat) - len_before:
print(f'{(len_before - len(df_data_vax_syms_flat)):>10} duplicates dropped in df_data_vax_syms_flat') ; print()
'''
SMALLER SYMPTOM_TEXT, REMOVAL OF REPEAT SENTENCES, DEDUPE
'''
df_data_vax_syms_flat = symptoms_dedupe_repeat_sentences(df_data_vax_syms_flat)
''' Save result into one file like ... 2020-12-25_VAERS_FLATTENED.csv '''
filename_flattened = dir_flattened + '/' + files_date_marker + '_VAERS_FLATTENED.csv'
print(f' Saving result into one file: {filename_flattened}') ; print()
write_to_csv(df_data_vax_syms_flat, filename_flattened)
print(f'{len(df_data_vax_syms_flat):>10} rows with unique VAERS_IDs in {filename_flattened}') ; print()
print(f' Flattening of {files_date_marker} done') ; print()
df_flat_2 = df_data_vax_syms_flat
dict_done_flag['flat2'] = files_date_marker
return
'''
Some VAERS_ID in 2021-01-07 for example with multiple lots/doses (multiple rows in VAX file) for testing
903999 903157 903848 903867 904522 905340 906428 907571 907776 907837 908431 908448 908763 909370 909520 910580 919620
18 doses: https://medalerts.org/vaersdb/findfield.php?IDNUMBER=1900339&WAYBACKHISTORY=ON '''
# Integrated from original: compare()
def compare(date_currently):
'''
Compare based on flattened (one row per VAERS_ID with each row containing all of the information)
saving modifications to changes column.
Add reports to result set when not already there.
VAX file columns:
VAERS_ID VAX_TYPE VAX_MANU VAX_LOT VAX_DOSE_SERIES VAX_ROUTE VAX_SITE VAX_NAME
There are multiple entries with same VAERS_ID, different VAX_LOT and VAX_DOSE_SERIES '''
global df_flat_1
if not covid_earliest_vaers_id:
return
files_populate_information()
file_flattened_previous = get_flattened('previous', date_currently) # like ... 2022-10-28_VAERS_FLATTENED.csv
file_flattened_working = get_flattened('working' , date_currently) # like ... 2022-11-04_VAERS_FLATTENED.csv
if not file_flattened_previous:
print() ; print(f' No flattened file to compare prior to this, {date_currently}, skip compare')
file_for_copy_original = dir_compared + '/' + date_currently + '_VAERS_FLATFILE.csv'
print(f' Due to first drop, copying current {file_flattened_working} to: {file_for_copy_original}') ; print()
shutil.copy(file_flattened_working, file_for_copy_original)
# Give it these columns for consistency
df_flat_init = open_file_to_df(file_for_copy_original, doprint=0)
df_flat_init['cell_edits'] = 0
df_flat_init['status' ] = ''
df_flat_init['changes' ] = ''
df_flat_init = set_columns_order(df_flat_init)
write_to_csv(df_flat_init, file_for_copy_original, open_file=0)
# Initial row in stats file when only one compare/changes file
do_never_ever( df_flat_init.VAERS_ID.to_list(), date_currently, 'compare on df_flat_init')
do_ever_covid( sorted(set(df_flat_init.VAERS_ID.to_list())) )
stats_resolve(date_currently)
return
previous_date = date_from_filename(file_flattened_previous)
if not previous_date: # first run?
exit('No previous_date, cannot continue')
''' COMPARED '''
if len(df_flat_1):
print(f" Using flat {dict_done_flag['flat1']} already in memory, {len(df_flat_1)} rows")
df_flat_prv = df_flat_1.copy() # the one finished previously, carryover
else:
df_flat_prv = open_file_to_df(file_flattened_previous, doprint=1)
check_dupe_vaers_id(df_flat_prv)
''' FLATTENED '''
if len(df_flat_2): # global from def flattening
print(f" Using flat {dict_done_flag['flat2']} already in memory, {len(df_flat_2)} rows") ; print()
df_flat_new = df_flat_2.copy() # the one just finished in the flattening (not compare or changes), carryover
df_flat_new = types_set(df_flat_new)
else:
df_flat_new = open_file_to_df(file_flattened_working, doprint=1)
check_dupe_vaers_id(df_flat_new)
if not stats['drop_input_covid']:
stats['drop_input_covid'] = len(df_flat_new)
print(f"=== Added count_input {stats['drop_input_covid']} in def compare")
dict_done_flag['flat1'] = date_currently
''' cell_edits and changes fields only exist in the changes file
Have to be pulled from there to be updated when applicable '''
print() ; print(f' Previous changes file for changes, cell_edits and status columns')
''' Previous changes filename, conditions, for compare '''
# KEEP FOR NOW if len(df_flat_prv):
# KEEP FOR NOW print(f" Using changes {dict_done_flag['changes']} already in memory")
# KEEP FOR NOW elif len( files['changes']['date'] ):
# KEEP FOR NOW dates_changes = files['changes']['date']
# KEEP FOR NOW date_prv_candidates = [x for x in dates_changes if x < date_currently]
# KEEP FOR NOW date_changes_prv = date_prv_candidates[-1] # most recent
# KEEP FOR NOW filename_changes_previous = files['changes']['keyval'][date_changes_prv] # like TODO
# KEEP FOR NOW df_flat_prv = open_file_to_df(filename_changes_previous, doprint=1)
# KEEP FOR NOW del dates_changes, date_prv_candidates, date_changes_prv, filename_changes_previous
# KEEP FOR NOW
# KEEP FOR NOW else:
# KEEP FOR NOW ''' Resort to initializing with flattened. '''
# KEEP FOR NOW print(f' Due to first CHANGES, using {file_flattened_previous}')
# If first run, it needs these columns since only changes files contain these columns
if 'cell_edits' not in df_flat_prv.columns: df_flat_prv['cell_edits'] = int(0)
if 'changes' not in df_flat_prv.columns: df_flat_prv['changes' ] = ''
if 'status' not in df_flat_prv.columns: df_flat_prv['status' ] = ''
df_flat_prv = df_flat_prv.sort_values(by=['cell_edits', 'status', 'changes'], ascending=False)
if nan_alert(df_flat_prv):
pause=1 # nans in ['AGE_YRS', 'CAGE_YR', 'CAGE_MO', 'HOSPDAYS', 'NUMDAYS']
del file_flattened_previous, file_flattened_working
#df_flat_prv_ori = df_flat_prv.copy() ; df_flat_new_ori = df_flat_new.copy()
print()
print(f'{" "*30} Comparing')
print() ; print(f'{" "*30} {previous_date} v. {date_currently}')
'''
1. Identical in both prv and new
2. Excess in prv
a. Deleted in new
b.
3. Excess in new
a. Brand new above highest prv ID
b. Restored
c.
4. Both prv and new remaining
a. Changed
'''
# tmp for testing, normally done in flat but using flat files cooked up manually
if use_test_cases:
df_flat_new = symptoms_dedupe_repeat_sentences(df_flat_new)
df_flat_prv = symptoms_dedupe_repeat_sentences(df_flat_prv)
print()
print(f'{ len(df_flat_new):>10} this drop total covid')
print(f'{ len(df_flat_prv):>10} previous total covid')
''' Initialize df_edits, starting with df_flat_new,
then they will be changed in some cases later, eventually going to df_changes_done '''
# for ensuring all present on return
df_both_flat_inputs = pd.concat([df_flat_prv.reset_index(drop=True), df_flat_new.reset_index(drop=True) ], ignore_index=True).drop_duplicates(subset='VAERS_ID')
df_edits = df_flat_new.copy()
df_changes_done = pd.DataFrame(columns=list(df_flat_prv.columns))
list_vids_changes_in_edits = df_flat_prv.loc[ df_flat_prv.VAERS_ID.isin(df_edits .VAERS_ID) ].VAERS_ID.to_list()
list_vids_edits_in_prv = df_edits.loc [ df_edits .VAERS_ID.isin(df_flat_prv.VAERS_ID) ].VAERS_ID.to_list()
list_vids_in_both = list( set(list_vids_changes_in_edits) & set(list_vids_edits_in_prv) )
print(f'{len(list_vids_in_both):>10} in both previous and new, copying their values in edits, status, changes')
# Bring in the previous changes etc
list_vids_both_only_to_copy = sorted( set(df_flat_prv.VAERS_ID.to_list()) & set(df_edits.VAERS_ID.to_list()) )
list_vids_w_changes_in_both = df_flat_prv.loc[df_flat_prv.cell_edits.ne(0) | df_flat_prv.changes.ne('') | df_flat_prv.status.ne('')].VAERS_ID.to_list()
# Only those with meaningful (changed values) to copy
list_vids_filtered_both_only_to_copy = sorted( set(list_vids_both_only_to_copy) & set(list_vids_w_changes_in_both) )
if len(list_vids_w_changes_in_both):
df_edits = copy_column_from_1_to_2_per_vids('cell_edits', df_flat_prv, df_edits, list_vids_filtered_both_only_to_copy)
df_edits = copy_column_from_1_to_2_per_vids('changes' , df_flat_prv, df_edits, list_vids_filtered_both_only_to_copy)
df_edits = copy_column_from_1_to_2_per_vids('status' , df_flat_prv, df_edits, list_vids_filtered_both_only_to_copy)
df_edits = df_edits.copy().sort_values(by=['cell_edits', 'status', 'changes'], ascending=False)
# Ensure types are set correctly regardless of whether there are changes
df_edits = types_set(df_edits)
warn_mixed_types(df_edits)
if nan_alert(df_edits):
pause=1 # for breakpoint in debugger
# Add any others from df_flat_prv not in df_edits
df_flat_prv_not_in_edits = df_flat_prv.loc[ ~df_flat_prv.VAERS_ID.isin(df_edits.VAERS_ID) ] # PICKS UP DELETED ALSO
print(f' Concat of {len(df_flat_prv_not_in_edits)} previous that were not in edits')
if len(df_flat_prv_not_in_edits):
df_edits = pd.concat([ df_edits.reset_index(drop=True), df_flat_prv_not_in_edits.reset_index(drop=True) ], ignore_index=True)
# After concat, types may get mixed up, so reset them
df_edits = types_set(df_edits)
''' Identical. prv and new
Set cell_edits 0 and nothing in changes column or status.
Move to df_changes_done (removing from df_flat_prv and df_flat_new) '''
print(f' Moving identical rows to done')
df_merged = df_flat_prv.astype(str).fillna('').merge(df_flat_new.astype(str).fillna(''), indicator=True, how='outer', suffixes=('1', '2',))
# note the above made everything str df_merged = types_set(df_merged) # VAERS_ID back to str
df_merged = df_merged.sort_values(by=[ '_merge', 'cell_edits' ], ascending=False)
df_merged = move_column_forward(df_merged, '_merge') # comparing them
df_identical = df_merged.loc[ df_merged._merge.eq('both') ] # adding these to done
del df_identical['_merge']
vids_identical = df_identical.VAERS_ID.astype(int).to_list() # .astype(int) critical
# Move identicals to done, also remove these IDs in prv and new
if vids_identical:
df_edits, df_changes_done = move_rows(df_edits.loc[ df_edits.VAERS_ID.isin(vids_identical) ], df_edits, df_changes_done)
print(f' Removing identicals done from previous and new')
df_flat_prv = df_flat_prv.loc[ ~df_flat_prv.VAERS_ID.isin(vids_identical) ]
df_flat_new = df_flat_new.loc[ ~df_flat_new.VAERS_ID.isin(vids_identical) ]
print(f'{len(vids_identical):>10} identical set aside into df_changes_done (first content)')
######del df_merged, df_identical, vids_identical
print(f'{len(df_flat_new):>10} this drop remaining to work with')
print(f'{len(df_flat_prv):>10} previous remaining to work with')
print(f'{len(df_flat_new) - len(df_flat_prv):>10} difference')
# New stuff can be one of three things: higher than the highest prv VAERS_ID or gapfill or the restore of a deleted report
list_new_not_in_prv = df_flat_new.loc[ ~df_flat_new.VAERS_ID.isin(df_flat_prv.VAERS_ID) ].VAERS_ID.to_list()
''' Deleted. Flat prv-only
897309 NOT DELETED 897309 42 Deleted 2020-12-25 0 U USPFIZER INC2020443518 2020-11-17 2020-11-18 probably polycellulitis; The whole half of the arm was really red and swollen; The whole half of the arm was really red and swollen; This is a spontaneous report from a contactable pharmacist. A patient of unspecified age and gender received pneumococcal 13-val conj vac (dipht crm197 protein) (PREVNAR 13), intramuscular on an unspecified date at single dose for immunization. The patient's medical history and concomitant medications were not reported. The patient experienced polycellulitis, the whole half of the arm was really red and swollen on an unspecified date. The reporter mentioned that a patient developed something that the reporter thought was probably polycellulitis after shot of unknown pneumonia vaccine product; product might have been Prevnar 13 but reporter was not sure. The patient called the doctor who sent them to a clinic to be seen that day and think theirs was maybe 3 days since they had gotten the unknown pneumonia vaccine if the reporter remembered correctly. This occurred maybe 6 months ago. The reporter just remembered the whole half of the arm was really red and swollen. The outcome of the events was unknown. Lot/batch number has been requested.; Sender's Comments: A contributory role of PREVNAR 13 cannot be fully excluded in triggering the onset of probably polycellulitis with the whole half of the arm was really red and swollen. The impact of this report on the benefit/risk profile of the Pfizer product is evaluated as part of Pfizer procedures for safety evaluation, including the review and analysis of aggregate data for adverse events. Any safety concern identified as part of this review, as well as any appropriate action in response, will be promptly notified to Regulatory Authorities, Ethics Committees and Investigators, as appropriate. PNC13 Pfizer-Wyeth UNK OT PNEUMO PREVNAR13 0 0 0 U 0 UNK 2 _|_Cellulitis_|_Erythema_|_Peripheral swelling_|_
They all exist in df_edits but not in df_flat_new.
Add cell_edits +42 and in status column add Deleted.
Move to df_changes_done, removing from df_edits and df_flat_prv, also from df_flat_new as no compare possible of course. '''
list_deleted_prior = []
list_prv_not_in_new = df_flat_prv.loc[ ~df_flat_prv.VAERS_ID.isin(df_flat_new.VAERS_ID) ].VAERS_ID.to_list()
if len(list_prv_not_in_new): # 12-25 deleted for real, 3 of them: [902986, 903003, 903130]
# Deleted already noted earlier, hangers-on
df_deleted_already_noted = df_edits.loc[ df_edits.VAERS_ID.isin(list_prv_not_in_new) & df_edits.status.str.contains(r'Deleted \d{4}-\d{2}-\d{2} $', na=False) ] # note end of line, $
list_deleted_prior = df_deleted_already_noted.VAERS_ID.to_list()
# Deleted new, not already noted earlier
df_deleted_to_notate_new = df_edits.loc[ df_edits.VAERS_ID.isin(list_prv_not_in_new) & ~df_edits.VAERS_ID.isin(df_deleted_already_noted.VAERS_ID) ]
list_deleted_to_notate_new = df_deleted_to_notate_new.VAERS_ID.to_list()
if len(list_deleted_to_notate_new):
df_edits.loc[ df_edits.VAERS_ID.isin(list_deleted_to_notate_new), 'status' ] += f'Deleted {date_currently} '
df_edits.loc[ df_edits.VAERS_ID.isin(list_deleted_to_notate_new), 'cell_edits' ] += len(columns_vaers)
len_newly_deleted = len(list_deleted_to_notate_new)
print(f'{ len_newly_deleted:>10} newly deleted kept: {subrange(list_deleted_to_notate_new, 6)}')
print(f'{len(list_deleted_prior):>10} deleted that were noted already: {subrange(list_deleted_prior, 6)}')
#df_edits, df_changes_done = move_rows(df_edits.loc[df_edits.VAERS_ID.isin(list_deleted_prior)], df_edits, df_changes_done)
#df_edits, df_changes_done = move_rows(df_edits.loc[df_edits.VAERS_ID.isin(list_deleted_to_notate_new)], df_edits, df_changes_done)
#df_flat_prv = df_flat_prv .loc[ ~df_flat_prv .VAERS_ID.isin(list_prv_not_in_new) ]
#df_flat_prv = df_flat_prv .loc[ ~df_flat_prv .VAERS_ID.isin(list_deleted_prior) ] # remove them from flat_prv, and they are already not in new
#df_flat_prv = df_flat_prv .loc[ ~df_flat_prv .VAERS_ID.isin(list_deleted_to_notate_new) ]
stats['deleted'] += len_newly_deleted
# These are in prv and NOT new so no compare is possible, they're done
df_edits, df_changes_done = move_rows(df_edits.loc[df_edits.VAERS_ID.isin(list_prv_not_in_new)], df_edits, df_changes_done)
df_flat_prv = df_flat_prv.loc[ ~df_flat_prv.VAERS_ID.isin(list_prv_not_in_new) ] # remove from prv
######del df_deleted_to_notate_new, list_deleted_to_notate_new
'''
1. Identical in both prv and new
2. Excess in prv
a. Deleted in new
b.
3. Excess in new
a. Brand new above highest prv ID
b. Restored
c. Gapfill
4. Both prv and new remaining
a. Changed
'''
''' Restored. Flat new that were Deleted but now being Restored
They all exist in df_edits and are marked Deleted.
Add cell_edits +42 and add status column Restored.
Remove them from df_flat_new but keep them in df_edits since there could be other changes also. '''
df_restored = df_edits.loc[ df_edits.VAERS_ID.isin(df_flat_new.VAERS_ID) & df_edits.status.str.contains(r'Deleted \d{4}-\d{2}-\d{2} $', na=False) ]
list_restored = df_restored.VAERS_ID.to_list()
if list_restored:
# WAIT, ARE THESE IN DF_EDITS OR JUST DF_FLAT_NEW?
df_edits.loc[ df_edits.VAERS_ID.isin(list_restored), 'cell_edits' ] += len(columns_vaers)
df_edits.loc[ df_edits.VAERS_ID.isin(list_restored), 'status' ] += f'Restored {date_currently} ' # Mark them as Restored
df_edits, df_changes_done = move_rows(df_edits.loc[ df_edits.VAERS_ID.isin(list_restored) ], df_edits, df_changes_done) # TODO: If changes also, those are missed here
###### WTF Do rerun #####df_flat_new = df_flat_new.loc[ ~df_flat_new.VAERS_ID.isin(list_restored) ] # remove them from df_flat_new being scrutinized
df_flat_prv = df_flat_prv.loc[ ~df_flat_prv.VAERS_ID.isin(list_restored) ] # some confusion, just removing, TODO
stats['restored'] += len(list_restored)
######del df_deleted_already_noted
''' Greater-Than. Flat new-only high VAERS_ID: df_flat_new VAERS_ID above max df_flat_prv
Set cell_edits 0 and nothing in changes or status column.
Move to df_changes_done (removing from df_flat_new) '''
list_flat_new_gt_vids = df_flat_new.loc[ df_flat_new.VAERS_ID.gt( df_flat_prv.VAERS_ID.max() ) ].VAERS_ID.to_list()
list_flat_new_Lt_vids = df_flat_new.loc[ df_flat_new.VAERS_ID.lt( df_flat_prv.VAERS_ID.max() ) ].VAERS_ID.to_list()
''' Gapfill/Throttled/Delayed/Late. Flat new-only remaining. (VAERS_IDs that didn't show up in in the sequence their drop but only later)
First appearance, late. Is possible because all greater-than prv.max() have already been moved.
Move from df_edits to df_changes_done and remove from df_flat_new '''
list_prv_vids = df_flat_prv.VAERS_ID.to_list()
list_gapfills = sorted( set(list_flat_new_Lt_vids) - set(list_prv_vids) )
list_gapfills = sorted( set(list_gapfills) - set(list_restored) ) # cludgy but seems to get the job done
if len(list_gapfills):
df_edits = df_edits.copy().sort_values(by=['status', 'changes', 'cell_edits'], ascending=False)
df_edits.loc[ df_edits.VAERS_ID.isin(list_gapfills), 'status' ] += f'Delayed {date_currently} ' # staying with the term Delayed for now
stats['gapfill'] += len(list_gapfills)
# These are in new and NOT prv so no compare is possible, they're done
df_edits, df_changes_done = move_rows(df_edits.loc[df_edits.VAERS_ID.isin(list_gapfills)], df_edits, df_changes_done)
df_flat_new = df_flat_new.loc[ ~df_flat_new.VAERS_ID.isin(list_gapfills) ] # remove from new
check_dupe_vaers_id(df_edits)
check_dupe_vaers_id(df_changes_done)
print(f'{ len(list_flat_new_gt_vids):>10} new (higher than max VAERS_ID in previous) for {date_currently}')
print(f'{ len(list_gapfills):>10} delayed this drop, filling gaps' )
print(f'{ len_newly_deleted:>10} newly deleted this drop kept' )
print(f'{ len(list_restored):>10} restored this drop {subrange(list_restored, 6)}' )
######del list_flat_new_gt_vids, list_gapfills, list_prv_not_in_new, list_restored, len_newly_deleted
len_before = len(df_flat_prv)
df_flat_prv = df_flat_prv.drop_duplicates(subset=df_flat_prv.columns, keep='last')
if len_before - len(df_flat_prv): # Should be none. Overwrite of values now done
print(f'\n\n\n\n\t\t\t\t {(len_before - len(df_flat_prv)):>10} complete duplicates dropped in df_flat_prv, SHOULD NOT HAPPEN \n\n\n\n')
len_before = len(df_flat_new)
df_flat_new = df_flat_new.drop_duplicates(subset=df_flat_new.columns, keep='last') # Should be none. Overwrite of values now done
if len_before - len(df_flat_new):
print(f'\n\n\n\n\t\t\t\t {(len_before - len(df_flat_new)):>10} complete duplicates dropped in df_flat_new, SHOULD NOT HAPPEN \n\n\n\n')
# Brand new reports only to done, no compare needed
df_only_flat_new = df_flat_new.loc[ ~df_flat_new.VAERS_ID.isin(df_flat_prv.VAERS_ID) ]
list_only_flat_new = df_only_flat_new.VAERS_ID.to_list()
if list_only_flat_new:
df_flat_new = df_flat_new.loc[ ~df_flat_new.VAERS_ID.isin(list_only_flat_new) ]
df_edits, df_changes_done = move_rows(df_edits.loc[df_edits.VAERS_ID.isin(list_only_flat_new)], df_edits, df_changes_done)
# Ensure prv and new are now the same reports, to be compared
if prv_new_error_check(df_flat_prv, df_flat_new):
pause=1 # nans in ['CAGE_YR', 'CAGE_MO', 'HOSPDAYS']
if nan_alert(df_flat_prv) or nan_alert(df_flat_new):
pause=1
''' Remaining are only common VAERS_IDs and same number of rows containing some fields that vary. '''
df_flat_prv = df_flat_prv.sort_values(by='VAERS_ID') ; df_flat_prv = df_flat_prv.reset_index(drop=True)
df_flat_new = df_flat_new.sort_values(by='VAERS_ID') ; df_flat_new = df_flat_new.reset_index(drop=True)
df_flat_new = df_flat_new.reindex(df_flat_prv.columns, axis=1) # Same column order. TODO: Find out how the columns changed order after adding the blanks block of code below
if len(df_edits):
if nan_alert(df_edits):
pause=1
len_edits = 0 # ongoing count of modified reports
vid_cols_affected = {}
warn_mixed_types(df_edits)
df_flat_prv = types_set(df_flat_prv)
warn_mixed_types(df_flat_prv)
warn_mixed_types(df_flat_new)
warn_mixed_types(df_changes_done)
list_prv_and_new_both = set( df_flat_prv.VAERS_ID.to_list() ) & set( df_flat_new.VAERS_ID.to_list() )
if not len(df_flat_new):
print(f'{str(0):>10} column changes in {len(columns_vaers)} columns')
else:
print() ; print(' Column value changes') ; print() # section could do with some refactoring smarter surely
try:
# List of columns changed, skipping cell_edits, status and changes
df_all_changed = df_flat_prv.compare(df_flat_new) # TODO: Many nans
cols_changed_list = sorted( set( [ x[0] for x in list(df_all_changed.columns) ] ) )
except Exception as e: # note error, patch and allow to continue
error(f' prv and new len diff {e} {inspect.stack()[1][2]}')
print(f'{line()} {e}')
if len(df_flat_prv) > len(df_flat_new): # TODO: Is this insane????? WHY ARE THEY DIFFERENT IN THE FIRST PLACE????
df_flat_prv = df_flat_prv.loc[ df_flat_prv.VAERS_ID.isin(df_flat_new.VAERS_ID) ]
df_flat_prv = df_flat_prv.reset_index(drop=True)
df_flat_new = df_flat_new.reset_index(drop=True)
elif len(df_flat_prv) < len(df_flat_new):
df_flat_new = df_flat_new.loc[ df_flat_new.VAERS_ID.isin(df_flat_prv.VAERS_ID) ]
df_flat_prv = df_flat_prv.reset_index(drop=True)
df_flat_new = df_flat_new.reset_index(drop=True)
df_all_changed = df_flat_prv.compare(df_flat_new)
cols_changed_list = sorted( set( [ x[0] for x in list(df_all_changed.columns) ] ) )
cols_changed_list = [x for x in cols_changed_list if x not in ['cell_edits', 'status', 'changes']]
check_dupe_vaers_id(df_flat_prv)
check_dupe_vaers_id(df_flat_new)
######del df_all_changed
count_cols_altered = len(cols_changed_list)
show_col_row_progress = 0
count_cols = 0
''' Debug utility
Specify a VAERS_ID in vid_target
and/or a debug_cols to focus on. '''
debug_dates = [] #['2021-01-15'] # list of dates like '2021-01-15' for pause if any and if breakpoint set
debug_vids = [903806] # list of int VAERS_ID for pause if any and if breakpoint set
debug_cols = [] #['SPLTTYPE', 'ER_ED_VISIT'] # like 'SYMPTOM_TEXT' optional for debug
for d in debug_dates:
if date_currently == d:
print(f'\n\n\n\n\t\t\t breakpoint for date {d} \n\n\n\n')
blank_brackets = '[]'
# Bug TODO: The occasional ... [] <> [] ... is a bug, missing content on one side or the other.
for col in cols_changed_list: # Find the new changes
#if debug_cols: # debug
# if col not in debug_cols:
# continue
if use_test_cases and col == 'FORM_VERS': continue
count_cols += 1
col_padded = (' ' * (20 - len(col))) + col
df_slice_prv = df_flat_prv[['VAERS_ID', col]].sort_values(by='VAERS_ID')
df_slice_new = df_flat_new[['VAERS_ID', col]].sort_values(by='VAERS_ID')
df_slice_prv = df_slice_prv.reset_index(drop=True) # make these indexes same
df_slice_new = df_slice_new.reset_index(drop=True)
vids_changed_this_col = df_slice_new.loc[ df_slice_new[col].ne(df_slice_prv[col]) ].VAERS_ID.to_list()
# Just the changes
df_three_columns = pd.merge(
df_slice_prv.loc[df_slice_prv.VAERS_ID.isin(vids_changed_this_col)],
df_slice_new.loc[df_slice_new.VAERS_ID.isin(vids_changed_this_col)],
on='VAERS_ID', suffixes=('_prv', '_new'))
if nan_alert(df_three_columns):
pause=1
del df_slice_prv, df_slice_new
col_prv = col + '_prv'
col_new = col + '_new'
count_rows = 0
if debug_breakpoint(date_currently, col, df_three_columns.VAERS_ID.to_list(), debug_dates, debug_cols, debug_vids):
pause=1 # a line for setting a breakpoint if conditions match
#else:
# continue
len_before = len(df_three_columns)
check_dupe_vaers_id(df_three_columns)
# necessary? TODO: df_three_columns = df_three_columns.drop_duplicates(subset=df_three_columns.columns, keep=False)
show_dupe_count = 0 # no longer wanting to print this
if show_dupe_count and (len(df_three_columns) - len_before):
print(f'{(len_before - len(df_three_columns)):>10} duplicates dropped in df_three_columns (columns VAERS_ID, prv and new), should not happen')
''' Special case the 11-11 purge in 2022 to try to increase speed
and to reduce the size of run output vastly by marking them in bulk as blanked out
These bulk blankings in the purge are not counted in column changes: {} '''
df_3_made_blank = df_three_columns.loc[ df_three_columns[col_prv].ne('') & df_three_columns[col_new].eq('') ]
if len(df_3_made_blank) >= 200:
vids_list = df_3_made_blank.VAERS_ID.to_list()
df_blanked_all = df_edits.loc[ df_edits.VAERS_ID.isin(vids_list) ]
print(f' {len(df_3_made_blank):>10} {col} blanked out, noting in bulk, omitting their VAERS_IDs')
df_blanked_all = df_blanked_all.copy() # quirky requirement here, what's the point in having df one can't edit without making a copy first?
message = f"{len(df_3_made_blank)} rows with {df_3_made_blank[col_prv].astype(str).map(len).sum()} total bytes MADE BLANK"
print(f'{" ":>10} {col_padded} {" ":>8} {message:>70} <> [] ')
stats['cells_emptied'] += len(vids_list)
df_blanked_all['changes_old'] = df_blanked_all['changes']
# col_prv to 'changes_new', a new column
df_blanked_all.loc[df_blanked_all.VAERS_ID.isin(vids_list), 'changes_new'] = df_blanked_all['VAERS_ID'].map(
pd.Series(df_3_made_blank[col_prv].values, index=vids_list).to_dict())
if nan_alert(df_blanked_all):
pause=1
df_blanked_all['changes_a'] = f"{col} {date_currently}: " # section clumsy! Help!?
df_blanked_all['changes_b'] = f" <> [] "
df_blanked_all['changes'] = df_blanked_all['changes_old'] + df_blanked_all['changes_a'] + df_blanked_all['changes_new'] + df_blanked_all['changes_b']
del df_blanked_all['changes_old'] ; del df_blanked_all['changes_new'] ; del df_blanked_all['changes_a'] ; del df_blanked_all['changes_b']
df_blanked_all = df_blanked_all.reset_index(drop=True) # avoid ... cannot reindex from a duplicate axis ... in copy_column_from_1_to_2_per_vids()
df_edits = df_edits.reset_index(drop=True) # no idea why this suddenly became necessary
''' duplicate values in your original index. To find them do this:
df_blanked_all[df_blanked_all.index.duplicated()]
df_edits[df_edits.index.duplicated()] '''
df_edits.loc[ df_edits.VAERS_ID.isin(vids_list), 'cell_edits' ] += 1 # direct
df_edits = copy_column_from_1_to_2_per_vids('changes', df_blanked_all, df_edits, vids_list) # copied into
df_edits = copy_column_from_1_to_2_per_vids('status' , df_blanked_all, df_edits, vids_list)
df_three_columns = df_three_columns.loc[ ~df_three_columns.VAERS_ID.isin(vids_list) ] # remove them from df_three_columns
if nan_alert(df_changes_done):
pause=1
if not len(df_three_columns): # ok in this case
continue
if not len(df_three_columns):
print(f'\n\n\n\n\t\t\t df_three_columns HAS NO CHANGES, UNEXPECTED, SOMETHING IS WRONG \n\n\n\n')
continue
df_three_columns = df_three_columns.sort_values(by=[col_prv, col_new])
''' Remove same ignoring punctuation.
1. Often vastly reduces the work for diff_context() to do, like on 2021-09-03 from 496837 rows to 172.
2. Ignores for example commas changed to pipe like 1483307 on 2021-09-03 in SYMPTOM_TEXT, was otherwise annoying.
CDC went wild on 2021-09-03 for example, changing commas to pipe.
CDC returned back from pipe to commas on 2022-01-07, sweeping trivial changes, unless my files are corrupted.
Now keeping pipe in VAX_LOT etc but ignoring in other fields with inexplicable changes like ... ALLERGIES 1075898 fentanyl,codeine,steroid,fruits,nuts <> fentanyl|codeine|steroid|fruits|nuts
... change of plan:
Just treating DATEs differently, everything else the same, as not much info is lost, otherwise filled with trivial noise.
'''
# Ok, downright mysterious, fingers crossed it's correct
len_before = len(df_three_columns)
if 'DATE' in col: # neutralize hundreds of thousands of date changes dropping 0's like 12/03/2020 <> 12/3/2020
df_three_columns = df_three_columns.loc[df_three_columns[col_prv].astype(str).str.replace(r'^(?:0*\d\/\d\d|\d\d\/0*\d|0*\d\/0*\d)\/\d{4}', '', regex=True) != df_three_columns[col_new].astype(str).str.replace(r'^(?:0*\d\/\d\d|\d\d\/0*\d|0*\d\/0*\d)\/\d{4}', '', regex=True)]
else:
df_three_columns = df_three_columns.loc[df_three_columns[col_prv].astype(str).str.replace(r'[\W_]', '', regex=True) != df_three_columns[col_new].astype(str).str.replace(r'[\W_]', '', regex=True)]
''' Line above can result in no changes, they were only punctuation etc '''
count_trivial_changes = len_before - len(df_three_columns)
if count_trivial_changes:
cellword = single_plural(count_trivial_changes, 'cells') #' cell' if count_trivial_changes == 1 else 'cells'
print(f'{col_padded} {count_trivial_changes:>7} {cellword} of trivial non-letter differences ignored')
stats['trivial_changes_ignored'] += count_trivial_changes
if not len(df_three_columns): # because with trivials removed, they're often all identical so there's often nothing left to compare
continue
# Faster way, drop lambda?: https://stackoverflow.com/questions/63306888/general-groupby-in-python-pandas-fast-way
# seeing ... No objects to concatenate ... due to a nan in 1254218 AGE_YRS_prv
df_three_columns = df_three_columns.fillna('')
try:
df_uniq = df_three_columns.groupby([col_prv, col_new])[['VAERS_ID', col_prv, col_new]].transform(lambda x: ' '.join(x.astype(str).unique()))
except Exception as e:
print(f'df_uniq df_three_columns transform lambda {len(df_three_columns)} {df_three_columns} Exception {e}')
error(f' df_uniq df_three_columns transform lambda {e} {inspect.stack()[1][2]}')
len_before = len(df_uniq)
df_uniq = df_uniq.drop_duplicates(subset=df_uniq.columns)
count_dupes = len_before - len(df_uniq)
if count_dupes:
print(f"{count_dupes:>10} {single_plural(count_dupes, 'duplicates')} dropped in df_three_columns")
df_uniq = df_uniq.copy()
df_uniq['VAERS_IDs'] = df_uniq.VAERS_ID.str.split(' ')
df_uniq = df_uniq[['VAERS_IDs', col_prv, col_new]] # dropping VAERS_ID strings with spaces where multiple
df_uniq['len'] = df_uniq[col_new].apply(len)
df_uniq = df_uniq.sort_values(by=list(set(df_uniq.columns) - set(['VAERS_IDs']))) # for print
for index, row in df_uniq.iterrows():
count_rows += 1
if show_col_row_progress and (count_rows == 1) or (count_rows % 100 == 0) and (count_cols != count_cols_altered):
# needs work, is here expecting updated progress indicators while code is working hard
print(f'{col_padded:>20} {count_cols}/{count_cols_altered} columns {count_rows}/{len(df_uniq)} rows ', end='\r')
vids_list = row['VAERS_IDs'] # all with this same change, often just one
vids_list = sorted([int(x) for x in vids_list])
if debug_vids:
for dv in debug_vids:
if dv in vids_list:
vid_pause = 1
vids_list_to_print = ''
vids_len_to_print = ''
vid_to_print = ''
vid = vids_list[0] # only one needs to be processed, then distribute the result to all the vids
val_prv = row[col_prv] ## Y
val_new = row[col_new] ## empty str
#blank_brackets = '[]' if not val_prv else '' # has to be captured before the diff
#blank_brackets = '[]' if not val_new else ''
val_prv_ori = val_prv ## Y
val_new_ori = val_new ## empty str # before diff_context() might change them
newly_cut = 1 if val_prv and (not val_new) and ('cut_' not in val_prv) else 0 ## newly_cut=1
continuing_cut = val_prv if 'cut_' in val_prv and not val_new else '' ## continuing_cut=''
restored = ''
if 'cut_' in val_prv and val_new and val_prv.startswith(val_new): # and (not val_new) and ('cut_' not in val_prv) else 0 ## continuing_cut=''
restored = ' [restored]'
#if continuing_cut:
# continue # it's fine left alone like 'Y cut_2021-01-08 '
if newly_cut: # completely blanked out
stats['cells_emptied'] += len(vids_list)
if debug_vids or debug_cols:
for v in vids_list:
if v not in vid_cols_affected:
vid_cols_affected[v] = [col]
if col not in vid_cols_affected[v]:
vid_cols_affected[v].append(col)
if col in ['AGE_YRS', 'CAGE_YR', 'CAGE_MO', 'HOSPDAYS', 'NUMDAYS']:
val_new = re.sub(r'\.0$', '', str(val_new)) # 30.0 to plain 30
val_prv = re.sub(r'\.0$', '', str(val_prv))
if val_prv and val_new: # both have content, pare down to the diff
if re.sub(r'\W', '', val_prv) == re.sub(r'\W', '', val_new): # skip trivial, for example SYMPTOM_TEXT 1272751 40,3 <> 40|3 ... and ... SYMPTOM_TEXT 1946413 21:15|(at <> 21:15,(at
continue
# DIFF, vast
val_prv, val_new = diff_context(val_prv, val_new, col, vid, date_currently)
elif not val_prv and not val_new: # both empty (no diffs) since trivial differences were removed they can wind up the same now
continue
if not continuing_cut: # not the case of was cut, still empty
stats['cells_edited'] += len(vids_list) # this does not count trivial changes like punctuation but some are minor
stats['columns'][col] += len(vids_list)
else:
pause=1
if len(vids_list) == 1: # fill the values to be shown (printed to screen) depending on single or list/multiple
vid_to_print = vid
else:
vids_len_to_print = len(vids_list) if len(vids_list) > 1 else ''
vids_list_to_print = [int(x) for x in vids_list] # no need for all those string single quotes
val_prv = blank_brackets if not val_prv else val_prv # confusions here
val_new = blank_brackets if not val_new else val_new ## val_new=[]
''' 2115523 German character? Or replacement character ? https://stackoverflow.com/a/72656394/962391
'ADHD; Adverse reaction to antibiotics (Nausea and vomiting induced by K?vepening.); Allergy; Asthma; Migraine'
UnicodeEncodeError: 'charmap' codec can't encode character '\\ufffd' in position 86: character maps to <undefined>
Possibly happens in command prompt but not in pycharm debugger '''
# Added later. Say DIED blanked became 'Y cut_2021-01-02', then restored. Want its previous to show as blank.
#if val_new and 'cut_' in val_prv_ori: # test this TODO c o n f u s i o n s
# val_prv = "''"
try:
''' Due especially to 2022-11-11 clearing out of SYMPTOM_TEXT, LAB_DATA, HISTORY etc
for print during run only, abbreviating large content when just blanked out. '''
if ' cut_' in val_prv_ori:
if continuing_cut:
val_prv = val_prv_ori # like 'Y cut_2021-01-08 '
elif restored:
val_prv = "''" # empty string instead of empty brackets indicating restored from empty good or bad?
val_prv = re.sub(r' cut_.*$', '', val_prv) # just the pure original version of it
if not val_new and len(val_prv) > 100:
prv_excerpt = re.sub(r'^(.{1,40}.*?)\b.*', r'\1', val_prv) + f' ... ~{len(val_prv) - 40} more'
if not prv_excerpt or not val_new:
pause=1
print(f'{vids_len_to_print:>10} {col_padded} {vid_to_print:>8} {prv_excerpt:>70} <> {val_new}{restored} {vids_list_to_print}')
else:
if not val_prv_ori or not val_new_ori:
pause=1
if not continuing_cut: # don't repeatedly print those blanked
print(f'{vids_len_to_print:>10} {col_padded} {vid_to_print:>8} {val_prv:>70} <> {val_new}{restored} {vids_list_to_print}')
except Exception as e:
''' Mystery. Example first dose shows up as question mark but is actually unicode: https://medalerts.org/vaersdb/findfield.php?IDNUMBER=2128531&WAYBACKHISTORY=ON
Attempt using r'�', val_prv = re.sub(r'�', '_', val_prv) ... out of Search and Replace program, but bogus
Exception workaround, 'charmap' codec can't encode character '<backslash>ufffd' in position 108: character maps to <undefined> ... '''
val_prv = re.sub(r'[^\x00-\x7F]+', '_', val_prv) # unicode replacement character
val_new = re.sub(r'[^\x00-\x7F]+', '_', val_new)
print(f' Exception workaround, {e} ...')
error(f' Exception workaround, {e} {inspect.stack()[1][2]}')
if not val_prv or not val_new:
pause = 1
print(f'{vids_len_to_print:>10} {col_padded} {vid_to_print} {val_prv:>70} <> {val_new}{restored} {vids_list_to_print}')
for vid in vids_list:
if col == 'DIED':
pause = 1
# If cleared out, blanked, deleted with val_new_ori = '' then keep the previous value in that cell
edits_add = 0
val_to_keep = val_prv if ( len(val_prv) and not len(val_new_ori) ) else val_new_ori
the_changes_note = ''
if newly_cut: #val_new_ori == blank_brackets: # Keep completely-deleted values in place but tag them with date like ... Myocarditis in a 42 year old cut_2022-11-11
edits_add = 1
val_to_keep = f'{val_to_keep} cut_{date_currently} ' # preserving deleted field contents, noting its date cut/deleted
the_changes_note = f'{col} cut_{date_currently} '
elif continuing_cut:
val_to_keep = val_prv_ori # with its cut_date content preserved
else:
if val_new and 'cut_' in val_prv_ori: # restored
val_prv = re.sub(r' cut_.*$', '', val_prv) # just the pure original version of it
if val_prv == val_new: # A value being restored to original if equal.
val_prv = "''" # It had been blanked out, existed preserved with it `cut_` note, needs to look like '' <> Y
# Only set to [] for the changes note if there's actual content to show
# Skip creating a changes note if both values are empty (would create [] <> [])
if not val_prv and not val_new:
# Both empty, don't create a change note
edits_add = 0
the_changes_note = ''
else:
if not val_prv: val_prv = '[]'
if not val_new: val_new = '[]'
edits_add = 1
the_changes_note = f'{col} {date_currently}: {val_prv} <> {val_new} '
if edits_add:
df_edits.loc[ df_edits.VAERS_ID.eq(vid), 'cell_edits' ] += 1
if the_changes_note:
df_edits.loc[ df_edits.VAERS_ID.eq(vid), 'changes' ] += the_changes_note
if val_to_keep:
df_edits.loc[ df_edits.VAERS_ID.eq(vid), col ] = val_to_keep
# cell_edits +1 for each cell that changed (non-trivially) on each report
print()
print(f"{ count_cols_altered:>10} {single_plural(count_cols_altered, 'columns')} altered")
del count_cols_altered
'''
Around here ...
FutureWarning: Downcasting object dtype arrays on .fillna, .ffill, .bfill is deprecated and will change in a future version.
Call result.infer_objects(copy=False) instead.
To opt-in to the future behavior, set `pd.set_option('future.no_silent_downcasting', True)`
'''
df_edits_unique = df_edits.drop_duplicates(subset=columns_vaers)
len_edits += len(df_edits_unique)
print(f"{len_edits:>10} modified {single_plural(len_edits, 'reports')} on {date_currently}") ; print()
if nan_alert(df_changes_done): # should not be necessary, TODO
pause=1
#df_changes_done['cell_edits'] = df_changes_done['cell_edits'].astype('float64').astype(int)
''' Add everything, with changes now set, into df_changes_done '''
df_edits, df_changes_done = move_rows(df_edits, df_edits, df_changes_done) # moving entire df_edits to df_changes_done
check_dupe_vaers_id(df_edits)
''' VAERS_IDs are int here '''
len_before = len(df_changes_done) # All columns
df_changes_done = df_changes_done.drop_duplicates(subset=df_changes_done.columns, keep='last') # Should be none. Overwrite of values now done
if len_before - len(df_changes_done):
print(f'{(len_before - len(df_changes_done)):>10} complete duplicates dropped in df_changes_done')
len_before = len(df_changes_done) # Just VAERS_ID, error-checking to make sure
df_changes_done = df_changes_done.drop_duplicates(subset='VAERS_ID', keep='last') # Should be none.
if len_before - len(df_changes_done):
print(f'\n\n\n\n\t\t\t\t{(len_before - len(df_changes_done)):>10} duplicate VAERS_ID dropped in df_changes_done, should never happen\n\n\n\n')
''' Sort the output by cell_edits. Section needs scrutiny.
'''
df_changes_done = df_changes_done.fillna('')
''' Annoying future warning and I don't know how best to resolve it. FutureWarning: Downcasting object dtype arrays on .fillna, .ffill, .bfill is deprecated and will change in a future version. Call result.infer_objects(copy=False) instead. To opt-in to the future behavior, set `pd.set_option('future.no_silent_downcasting', True)`
https://stackoverflow.com/questions/77900971/pandas-futurewarning-downcasting-object-dtype-arrays-on-fillna-ffill-bfill
'''
check_dupe_vaers_id(df_changes_done)
df_changes_done = df_changes_done.sort_values(by=['cell_edits', 'status', 'changes'], ascending=False)
if nan_alert(df_changes_done):
pause=1
''' VAERS_IDs with the most records/doses/lots in each report and at least one covid19.
(There's surely a better way with lambda or .apply() and/or .groupby() etc).
Here, just counting delimiters in VAX_LOT and adding 1 to that.
A report in df_vax has 18 entries in 2020-12-25 but no covid, for example, see df_vax.VAERS_ID.value_counts().unique()
vid_top_doses_list = sorted(df_doses.loc[ df_doses.doses.eq(df_doses.doses.max()) ].VAERS_ID.to_list())
TypeError: '<' not supported between instances of 'str' and 'int'
Tried df_changes_done.fillna('') above, didn't work.
Trying VAX_LOT.astype(str) <== TODO: Should be done earlier. Some must have been made in automatically when read.
'''
df_changes_done['VAX_LOT'] = df_changes_done.VAX_LOT.astype(str)
df_doses = df_changes_done.loc[df_changes_done.VAX_LOT.astype(str).str.contains(r'\|', na=False)][['VAERS_ID', 'VAX_LOT']]
if nan_alert(df_doses):
pause=1
df_doses['doses'] = df_doses.VAX_LOT.astype(str).str.count(r'\|')
filename = dir_compared + '/' + date_currently + '_VAERS_FLATFILE.csv'
print(f' Writing ... {filename}') ; print()
# When running against multiple input dates, open just the last one
do_open = 0
if files['input']['date'][-1] in filename: # Last one
do_open = 1
df_changes_done = set_columns_order(df_changes_done)
print()
print(f' This drop {do_elapsed(elapsed_drop)}')
print(f' Overall {do_elapsed(elapsed_begin)}')
print()
write_to_csv(df_changes_done, filename, open_file=do_open) # Has to be written here, the one big file. Separation of A and B later when necessary for the Excel rows limit.
# Thru this transition for clarity, df_changes_done becomes the next df_flat_prv (when processing multiple drops)
df_flat_1 = df_changes_done.copy() # use as the previous in next when processing more than one drop.
dict_done_flag[ 'changes' ] = date_currently # bookkeeping
#lookup(previous_date, date_currently, vid_cols_affected) # for debug, open a csv showing all of the reports touched for these dates compared
vid_top_doses_list = sorted(df_doses.loc[ df_doses.doses.eq(df_doses.doses.max()) ].VAERS_ID.to_list())
top_num = df_doses.doses.max() + 1
print(f'{len(vid_top_doses_list):>10} {single_plural(len(vid_top_doses_list), "reports")} with the most ({top_num}) records/lots/doses: {" ".join([str(x) for x in vid_top_doses_list])}')
#print(f"{stats['comparisons']:>10} {single_plural(stats['comparisons'], 'comparisons')} done")
vids_present = df_both_flat_inputs.VAERS_ID.to_list()
do_never_ever( vids_present, date_currently, 'compare on df_both_flat_inputs' )
do_ever_covid( vids_present )
stats_resolve( date_currently )
''' Ensure nothing is lost '''
verify_all_reports_present(df_both_flat_inputs, df_changes_done) # Relies on collection of IDs, new, never, ever seen
return
# Integrated from original: open_files()
def open_files(_date): # like './vaers_drop_inputs/2020-12-25'
''' Input files in dir_input:
csv within directories
zip files in a single directory, containing csv, treated as if they are folders, sort of. '''
files_populate_information()
if _date in files['consolidated']['date']: # already consolidated
print(f' {_date} already consolidated, no need to copy input files to dir_working')
shutil.rmtree(dir_working) # removing directory
os.mkdir(dir_working)
set_files_date_marker(dir_working, _date) # a flag used by consolidate()
return True # Already consolidated, so success
if _date in files['flattened']['date']:
print(f' Skipping unzip because flattened for {_date} already exists')
return True # Already processed, so success
if not _date in files['input']['keyval']:
exit(f" Failed to find in files['input']['keyval'] the _date {_date} in open_files() ")
files_value = files['input']['keyval'][_date]
if 'csv' in files_value:
print(f' Copy all {_date} to {dir_working}')
to_copy = [x for x in files['input']['files'] if _date in x]
shutil.rmtree(dir_working)
os.mkdir(dir_working)
for x in to_copy:
shutil.copy(x, dir_working)
elif isinstance(files_value, list): # another case earlier in development, csv files already extracted manually
print(f' Copy {_date}/* to {dir_working}')
shutil.rmtree(dir_working) # removing directory to avoid error next line
shutil.copytree(_date, dir_working)
set_files_date_marker(dir_working, _date)
elif 'zip' in files_value: # zip file, treat it sort of like a directory here
shutil.rmtree(dir_working)
os.makedirs(dir_working)
success = files_from_zip(files_value, dir_working)
if not success:
print(f' Skipping date {_date} due to zip file error')
return False # Indicate this date should be skipped
else:
exit(f' Unexpected _date {_date} in open_files() ')
set_files_date_marker(dir_working, _date) # a visual aid showing input file date marker, not really a file so much
return True # Indicate success
# Integrated from original: verify_all_reports_present()
def verify_all_reports_present(df_compare_input, df_compare_output): # Ensure nothing is lost
list_in_input_not_in_output = df_compare_input.loc[ ~df_compare_input.VAERS_ID.isin(df_compare_output.VAERS_ID) ].VAERS_ID.to_list()
if list_in_input_not_in_output:
print(f'\n\n\nERROR:\n{len(list_in_input_not_in_output)} VAERS_IDs in df_compare_input NOT in df_compare_output \n {subrange(list_in_input_not_in_output, 6)} \n\n')
set_vids_in_output = set( df_compare_output.VAERS_ID.to_list() )
set_ever_covid = set( ever_covid.keys() )
vids_in_df_not_in_ever_covid = set_vids_in_output - set_ever_covid
vids_not_in_df_in_ever_covid = set_ever_covid - set_vids_in_output
if vids_in_df_not_in_ever_covid:
print(f'{len(vids_in_df_not_in_ever_covid):>10} vids_in_df_not_in_ever_covid: {subrange(vids_in_df_not_in_ever_covid, 6)}') ; print()
if vids_not_in_df_in_ever_covid:
print(f'{len(vids_not_in_df_in_ever_covid):>10} vids_not_in_df_in_ever_covid: {subrange(vids_not_in_df_in_ever_covid, 6)}') ; print()
# Make certain none lost or extra
vids_set_of_all_in_prv_and_new = set( df_compare_input.VAERS_ID.to_list() )
vids_orphans = vids_set_of_all_in_prv_and_new - set(df_compare_output.VAERS_ID.to_list())
vids_extras = set(df_compare_output.VAERS_ID.to_list()) - vids_set_of_all_in_prv_and_new
if vids_orphans:
df_orphans = df_compare_input.loc[ ~df_compare_input.VAERS_ID.isin(df_compare_output.VAERS_ID) ]
print(f'\n\n\n{len(df_orphans):>10} orphans, records in df_compare_input that are not in df_compare_output')
print(' M U S T F I X \n\n\n')
write_to_csv(df_orphans, 'orphans.csv', open_file=1)
df_orphans, df_compare_output = move_rows(df_orphans, df_orphans, df_compare_output) # moving all of them out to done
if vids_extras:
df_extras = df_compare_output.loc[ ~df_compare_output.VAERS_ID.isin(df_compare_input.VAERS_ID) ]
print(f'\n\n\n{len(df_extras):>10} extras, records in df_compare_output that are not in df_compare_input')
print(f' M U S T F I X {subrange(df_extras.VAERS_ID.to_list(), 6)} \n\n\n')
write_to_csv(df_extras, 'extras.csv', open_file=1)
# Integrated from original: show_vid_as_text()
def show_vid_as_text(df, vid):
if not type(vid) is int: # TODO: Using type() instead of isinstance() for a typecheck. (unidiomatic-typecheck)
print(f' Expected vid {vid} to be an integer')
if vid not in df.VAERS_ID.to_list():
print(f' vid {vid} not found in the dataframe')
return
filename = f'{vid}.txt'
df = df.loc[ df.VAERS_ID.eq(vid) ]
df.to_csv(filename, sep=',', encoding='utf-8-sig', index=False)
sp.Popen(filename, shell=True)
# =============================================================================
# FINAL MERGED FILE CREATION
# =============================================================================
def create_final_merged_file():
"""
Create a final merged file from the latest FLATFILE in 2_vaers_full_compared.
This file contains all records with their complete change history.
"""
print()
print("="*80)
print("CREATING FINAL MERGED OUTPUT FILE")
print("="*80)
print()
# Find the latest FLATFILE
flatfiles = glob.glob(f'{dir_compared}/*_VAERS_FLATFILE.csv')
if not flatfiles:
flatfiles = glob.glob(f'{dir_compared}/*_VAERS_FLATFILE_A.csv')
if not flatfiles:
print(" No FLATFILE found in 2_vaers_full_compared directory")
return
# Sort by date in filename to get the latest
flatfiles.sort()
latest_file = flatfiles[-1]
print(f" Using latest file: {os.path.basename(latest_file)}")
try:
# Read the latest FLATFILE
print(f" Reading {os.path.basename(latest_file)}...")
df_final = pd.read_csv(latest_file, encoding='utf-8-sig', low_memory=False)
print(f" Total records: {len(df_final):,}")
# Create output filename with timestamp
output_filename = f'{dir_top}/VAERS_FINAL_MERGED.csv'
# Write the final merged file
print(f" Writing final merged file: {output_filename}")
write_to_csv(df_final, output_filename, open_file=False)
print()
print(f" ✓ Final merged file created successfully!")
print(f" Location: {output_filename}")
print(f" Records: {len(df_final):,}")
# Print summary statistics
if 'cell_edits' in df_final.columns:
total_edits = df_final['cell_edits'].sum()
records_with_edits = len(df_final[df_final['cell_edits'] > 0])
print(f" Total cell edits: {total_edits:,}")
print(f" Records with edits: {records_with_edits:,}")
if 'status' in df_final.columns:
deleted_count = len(df_final[df_final['status'].str.contains('Deleted', na=False)])
if deleted_count > 0:
print(f" Deleted records: {deleted_count:,}")
print()
except Exception as e:
error(f"Failed to create final merged file: {e}")
print(f" Error: {e}")
import traceback
traceback.print_exc()
# =============================================================================
# MAIN EXECUTION
# =============================================================================
def run_all():
"""Main execution function with all enhancements"""
print(__file__)
print()
print('='*80)
print('VAERS Enhanced Processing - Complete Edition')
print('='*80)
print()
print('Features:')
print(' ✓ Multi-core parallel processing')
print(' ✓ Memory-efficient chunked data handling')
print(' ✓ Command-line dataset selection (COVID/Full)')
print(' ✓ Progress bars for all operations')
print(' ✓ Comprehensive error tracking')
print(' ✓ Fixed stats functionality')
print(' ✓ All original processing logic integrated')
print()
# If merge-only flag is set, just create the final file and exit
if args.merge_only:
print('='*80)
print("MERGE-ONLY MODE: Creating final merged file")
print("="*80)
print()
create_final_merged_file()
return
if not validate_dirs_and_files():
exit_script("Directory validation failed")
print()
print("="*80)
print("STARTING PROCESSING")
print("="*80)
print()
# Main processing loop
while more_to_do():
this_drop_date = get_next_date()
print_date_banner(this_drop_date)
stats_initialize(this_drop_date)
# Try to open files, skip this date if it fails
if open_files(this_drop_date) == False:
print(f' Skipping processing for {this_drop_date} due to file errors')
files['done'].append(this_drop_date) # Mark as done to move to next
continue
consolidate(this_drop_date)
flatten(this_drop_date)
compare(this_drop_date)
# Create final merged output file
create_final_merged_file()
elapsed = do_elapsed(elapsed_begin)
print()
print("="*80)
print(f"PROCESSING COMPLETE - Total time: {elapsed}")
print("="*80)
print()
# Print final error summary
print_errors_summary()
if __name__ == "__main__":
try:
run_all()
except KeyboardInterrupt:
print("\n\nProcess interrupted by user")
exit_script()
except Exception as e:
error(f"Unexpected error in main: {type(e).__name__}: {e}")
import traceback
traceback.print_exc()
exit_script()